Structure-guided design of a potent Clostridiodes difficile toxin A inhibitor
- PMID: 36778856
- PMCID: PMC9909335
- DOI: 10.3389/fmicb.2023.1110541
Structure-guided design of a potent Clostridiodes difficile toxin A inhibitor
Erratum in
-
Corrigendum: Structure-guided design of a potent Clostridioides difficile toxin A inhibitor.Front Microbiol. 2023 Mar 30;14:1167817. doi: 10.3389/fmicb.2023.1167817. eCollection 2023. Front Microbiol. 2023. PMID: 37065148 Free PMC article.
Abstract
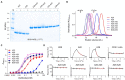
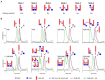
Crystal structures of camelid heavy-chain antibody variable domains (VHHs) bound to fragments of the combined repetitive oligopeptides domain of Clostridiodes difficile toxin A (TcdA) reveal that the C-terminus of VHH A20 was located 30 Å away from the N-terminus of VHH A26. Based on this observation, we generated a biparatopic fusion protein with A20 at the N-terminus, followed by a (GS)6 linker and A26 at the C-terminus. This A20-A26 fusion protein shows an improvement in binding affinity and a dramatic increase in TcdA neutralization potency (>330-fold [IC 50]; ≥2,700-fold [IC 99]) when compared to the unfused A20 and A26 VHHs. A20-A26 also shows much higher binding affinity and neutralization potency when compared to a series of control antibody constructs that include fusions of two A20 VHHs, fusions of two A26 VHHs, a biparatopic fusion with A26 at the N-terminus and A20 at the C-terminus (A26-A20), and actoxumab. In particular, A20-A26 displays a 310-fold (IC 50) to 29,000-fold (IC 99) higher neutralization potency than A26-A20. Size-exclusion chromatography-multiangle light scattering (SEC-MALS) analyses further reveal that A20-A26 binds to TcdA with 1:1 stoichiometry and simultaneous engagement of both A20 and A26 epitopes as expected based on the biparatopic design inspired by the crystal structures of TcdA bound to A20 and A26. In contrast, the control constructs show varied and heterogeneous binding modes. These results highlight the importance of molecular geometric constraints in generating highly potent antibody-based reagents capable of exploiting the simultaneous binding of more than one paratope to an antigen.
Keywords: Clostridiodes difficile; VHH; biparatopic; inhibitor; nanobody; single-domain antibody; toxin.
Copyright © 2023 Hussack, Rossotti, van Faassen, Murase, Eugenio, Schrag, Ng and Tanha.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Neutralization of Clostridium difficile toxin B with VHH-Fc fusions targeting the delivery and CROPs domains.PLoS One. 2018 Dec 12;13(12):e0208978. doi: 10.1371/journal.pone.0208978. eCollection 2018. PLoS One. 2018. PMID: 30540857 Free PMC article.
-
Neutralization of Clostridium difficile toxin A with single-domain antibodies targeting the cell receptor binding domain.J Biol Chem. 2011 Mar 18;286(11):8961-76. doi: 10.1074/jbc.M110.198754. Epub 2011 Jan 7. J Biol Chem. 2011. PMID: 21216961 Free PMC article.
-
Neutralizing epitopes on Clostridioides difficile toxin A revealed by the structures of two camelid VHH antibodies.Front Immunol. 2022 Nov 16;13:978858. doi: 10.3389/fimmu.2022.978858. eCollection 2022. Front Immunol. 2022. PMID: 36466927 Free PMC article.
-
Single domain camel antibodies: current status.J Biotechnol. 2001 Jun;74(4):277-302. doi: 10.1016/s1389-0352(01)00021-6. J Biotechnol. 2001. PMID: 11526908 Review.
-
Single domain Camelid antibody fragments for molecular imaging and therapy of cancer.Front Oncol. 2023 Sep 8;13:1257175. doi: 10.3389/fonc.2023.1257175. eCollection 2023. Front Oncol. 2023. PMID: 37746282 Free PMC article. Review.
Cited by
-
A review on camelid nanobodies with potential application in veterinary medicine.Vet Res Commun. 2024 Aug;48(4):2051-2068. doi: 10.1007/s11259-024-10432-x. Epub 2024 Jun 13. Vet Res Commun. 2024. PMID: 38869749 Review.
-
New treatment approaches for Clostridioides difficile infections: alternatives to antibiotics and fecal microbiota transplantation.Gut Microbes. 2024 Jan-Dec;16(1):2337312. doi: 10.1080/19490976.2024.2337312. Epub 2024 Apr 9. Gut Microbes. 2024. PMID: 38591915 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

