Gene differential co-expression analysis of male infertility patients based on statistical and machine learning methods
- PMID: 36778885
- PMCID: PMC9911419
- DOI: 10.3389/fmicb.2023.1092143
Gene differential co-expression analysis of male infertility patients based on statistical and machine learning methods
Abstract
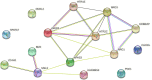
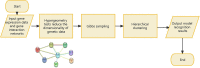
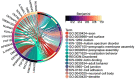
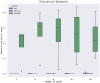
Male infertility has always been one of the important factors affecting the infertility of couples of gestational age. The reasons that affect male infertility includes living habits, hereditary factors, etc. Identifying the genetic causes of male infertility can help us understand the biology of male infertility, as well as the diagnosis of genetic testing and the determination of clinical treatment options. While current research has made significant progress in the genes that cause sperm defects in men, genetic studies of sperm content defects are still lacking. This article is based on a dataset of gene expression data on the X chromosome in patients with azoospermia, mild and severe oligospermia. Due to the difference in the degree of disease between patients and the possible difference in genetic causes, common classical clustering methods such as k-means, hierarchical clustering, etc. cannot effectively identify samples (realize simultaneous clustering of samples and features). In this paper, we use machine learning and various statistical methods such as hypergeometric distribution, Gibbs sampling, Fisher test, etc. and genes the interaction network for cluster analysis of gene expression data of male infertility patients has certain advantages compared with existing methods. The cluster results were identified by differential co-expression analysis of gene expression data in male infertility patients, and the model recognition clusters were analyzed by multiple gene enrichment methods, showing different degrees of enrichment in various enzyme activities, cancer, virus-related, ATP and ADP production, and other pathways. At the same time, as this paper is an unsupervised analysis of genetic factors of male infertility patients, we constructed a simulated data set, in which the clustering results have been determined, which can be used to measure the effect of discriminant model recognition. Through comparison, it finds that the proposed model has a better identification effect.
Keywords: Fisher test; Gibbs sampling; HPV; gene interaction network; hypergeometric distribution; machine learning; male infertility.
Copyright © 2023 Jia, Yin and Peng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aquila S., Sisci D., Gentile M., Middea E., Catalano S., Carpino A., et al. (2004). Estrogen receptor (ER) alpha and ER beta are both expressed in human ejaculated spermatozoa: evidence of their direct interaction with phosphatidylinositol-3-OH kinase/Akt pathway. J. Clin. Endocrinol. Metab. 89, 1443–1451. doi: 10.1210/jc.2003-031681, PMID: - DOI - PubMed
-
- Bollobás B., Borgs C., Chayes J. (2003). “Directed scale-free graphs,’’ in Proceedings of the fourteenth annual ACM-SIAM symposium on discrete algorithms. (Philadelphia, PA, USA). 132–139.
LinkOut - more resources
Full Text Sources

