Open-source, fully-automated hybrid cardiac substructure segmentation: development and optimisation
- PMID: 36780065
- PMCID: PMC10030448
- DOI: 10.1007/s13246-023-01231-w
Open-source, fully-automated hybrid cardiac substructure segmentation: development and optimisation
Abstract
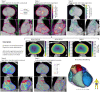
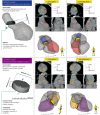
Radiotherapy for thoracic and breast tumours is associated with a range of cardiotoxicities. Emerging evidence suggests cardiac substructure doses may be more predictive of specific outcomes, however, quantitative data necessary to develop clinical planning constraints is lacking. Retrospective analysis of patient data is required, which relies on accurate segmentation of cardiac substructures. In this study, a novel model was designed to deliver reliable, accurate, and anatomically consistent segmentation of 18 cardiac substructures on computed tomography (CT) scans. Thirty manually contoured CT scans were included. The proposed multi-stage method leverages deep learning (DL), multi-atlas mapping, and geometric modelling to automatically segment the whole heart, cardiac chambers, great vessels, heart valves, coronary arteries, and conduction nodes. Segmentation performance was evaluated using the Dice similarity coefficient (DSC), mean distance to agreement (MDA), Hausdorff distance (HD), and volume ratio. Performance was reliable, with no errors observed and acceptable variation in accuracy between cases, including in challenging cases with imaging artefacts and atypical patient anatomy. The median DSC range was 0.81-0.93 for whole heart and cardiac chambers, 0.43-0.76 for great vessels and conduction nodes, and 0.22-0.53 for heart valves. For all structures the median MDA was below 6 mm, median HD ranged 7.7-19.7 mm, and median volume ratio was close to one (0.95-1.49) for all structures except the left main coronary artery (2.07). The fully automatic algorithm takes between 9 and 23 min per case. The proposed fully-automatic method accurately delineates cardiac substructures on radiotherapy planning CT scans. Robust and anatomically consistent segmentations, particularly for smaller structures, represents a major advantage of the proposed segmentation approach. The open-source software will facilitate more precise evaluation of cardiac doses and risks from available clinical datasets.
Keywords: Breast cancer; Cardiac substructures; Cardiotoxicity; Deep learning; Image segmentation; Lung cancer; Radiotherapy.
© 2023. Crown.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures







References
-
- Finnegan R, Lorenzen EL, Dowling J, Jensen I, Berg M, Thomsen MMS, et al. Analysis of cardiac substructure dose in a large, multi-centre Danish breast cancer cohort (the DBCG HYPO trial): Trends and predictive modelling. Radiother Oncol. 2020;153:130–8. doi: 10.1016/j.radonc.2020.09.004. - DOI - PubMed
-
- Iorio GC, Salvestrini V, Borghetti P, De Felice F, Greco C, Nardone V, et al. The impact of modern radiotherapy on radiation-induced late sequelae: focus on early-stage mediastinal classical Hodgkin Lymphoma. A critical review by the Young Group of the Italian Association of Radiotherapy and Clinical Oncology (AIRO) Crit Rev Oncol. 2021;161:103326. doi: 10.1016/j.critrevonc.2021.103326. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
