Four principles to establish a universal virus taxonomy
- PMID: 36780432
- PMCID: PMC9925010
- DOI: 10.1371/journal.pbio.3001922
Four principles to establish a universal virus taxonomy
Abstract
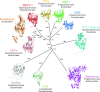
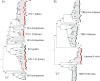
A universal taxonomy of viruses is essential for a comprehensive view of the virus world and for communicating the complicated evolutionary relationships among viruses. However, there are major differences in the conceptualisation and approaches to virus classification and nomenclature among virologists, clinicians, agronomists, and other interested parties. Here, we provide recommendations to guide the construction of a coherent and comprehensive virus taxonomy, based on expert scientific consensus. Firstly, assignments of viruses should be congruent with the best attainable reconstruction of their evolutionary histories, i.e., taxa should be monophyletic. This fundamental principle for classification of viruses is currently included in the International Committee on Taxonomy of Viruses (ICTV) code only for the rank of species. Secondly, phenotypic and ecological properties of viruses may inform, but not override, evolutionary relatedness in the placement of ranks. Thirdly, alternative classifications that consider phenotypic attributes, such as being vector-borne (e.g., "arboviruses"), infecting a certain type of host (e.g., "mycoviruses," "bacteriophages") or displaying specific pathogenicity (e.g., "human immunodeficiency viruses"), may serve important clinical and regulatory purposes but often create polyphyletic categories that do not reflect evolutionary relationships. Nevertheless, such classifications ought to be maintained if they serve the needs of specific communities or play a practical clinical or regulatory role. However, they should not be considered or called taxonomies. Finally, while an evolution-based framework enables viruses discovered by metagenomics to be incorporated into the ICTV taxonomy, there are essential requirements for quality control of the sequence data used for these assignments. Combined, these four principles will enable future development and expansion of virus taxonomy as the true evolutionary diversity of viruses becomes apparent.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Wildy P. Classifying viruses at higher levels: Symmetry and structure of virus particles as criteria. Symp Soc Gen Microbiol 1961;XII:145–63.

