An HLA-G/SPAG9/STAT3 axis promotes brain metastases
- PMID: 36780531
- PMCID: PMC9974476
- DOI: 10.1073/pnas.2205247120
An HLA-G/SPAG9/STAT3 axis promotes brain metastases
Abstract
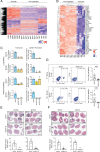
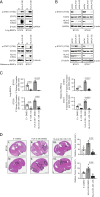
Brain metastases (BM) are the most common brain neoplasm in adults. Current BM therapies still offer limited efficacy and reduced survival outcomes, emphasizing the need for a better understanding of the disease. Herein, we analyzed the transcriptional profile of brain metastasis initiating cells (BMICs) at two distinct stages of the brain metastatic cascade-the "premetastatic" or early stage when they first colonize the brain and the established macrometastatic stage. RNA sequencing was used to obtain the transcriptional profiles of premetastatic and macrometastatic (non-premetastatic) lung, breast, and melanoma BMICs. We identified that lung, breast, and melanoma premetastatic BMICs share a common transcriptomic signature that is distinct from their non-premetastatic counterparts. Importantly, we show that premetastatic BMICs exhibit increased expression of HLA-G, which we further demonstrate functions in an HLA-G/SPAG9/STAT3 axis to promote the establishment of brain metastatic lesions. Our findings suggest that unraveling the molecular landscape of premetastatic BMICs allows for the identification of clinically relevant targets that can possibly inform the development of preventive and/or more efficacious BM therapies.
Keywords: HLA-G; SPAG9; STAT3; brain metastases.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

