AI reveals insights into link between CD33 and cognitive impairment in Alzheimer's Disease
- PMID: 36780558
- PMCID: PMC9956604
- DOI: 10.1371/journal.pcbi.1009894
AI reveals insights into link between CD33 and cognitive impairment in Alzheimer's Disease
Abstract
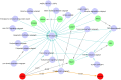
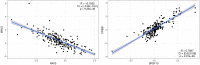
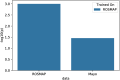
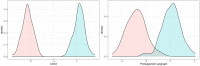
Modeling biological mechanisms is a key for disease understanding and drug-target identification. However, formulating quantitative models in the field of Alzheimer's Disease is challenged by a lack of detailed knowledge of relevant biochemical processes. Additionally, fitting differential equation systems usually requires time resolved data and the possibility to perform intervention experiments, which is difficult in neurological disorders. This work addresses these challenges by employing the recently published Variational Autoencoder Modular Bayesian Networks (VAMBN) method, which we here trained on combined clinical and patient level gene expression data while incorporating a disease focused knowledge graph. Our approach, called iVAMBN, resulted in a quantitative model that allowed us to simulate a down-expression of the putative drug target CD33, including potential impact on cognitive impairment and brain pathophysiology. Experimental validation demonstrated a high overlap of molecular mechanism predicted to be altered by CD33 perturbation with cell line data. Altogether, our modeling approach may help to select promising drug targets.
Copyright: © 2023 Raschka et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- P30 AG072975/AG/NIA NIH HHS/United States
- U01 AG046152/AG/NIA NIH HHS/United States
- RF1 AG057440/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- U01 AG061356/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- R01 NS080820/NS/NINDS NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- R01 AG018023/AG/NIA NIH HHS/United States
- U01 AG046170/AG/NIA NIH HHS/United States
- U24 AG061340/AG/NIA NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- R01 AG036836/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

