Genomic insights into antibiotic resistance and mobilome of lactic acid bacteria and bifidobacteria
- PMID: 36781180
- PMCID: PMC9930590
- DOI: 10.26508/lsa.202201637
Genomic insights into antibiotic resistance and mobilome of lactic acid bacteria and bifidobacteria
Abstract
Lactic acid bacteria (LAB) and Bifidobacterium sp. (bifidobacteria) can carry antimicrobial resistance genes (ARGs), yet data on resistance mechanisms in these bacteria are limited. The aim of our study was to identify the underlying genetic mechanisms of phenotypic resistance in 103 LAB and bifidobacteria using whole-genome sequencing. Sequencing data not only confirmed the presence of 36 acquired ARGs in genomes of 18 strains, but also revealed wide dissemination of intrinsic ARGs. The presence of acquired ARGs on known and novel mobile genetic elements raises the possibility of their horizontal spread. In addition, our data suggest that mutations may be a common mechanism of resistance. Several novel candidate resistance mechanisms were uncovered, providing a basis for further in vitro studies. Overall, 1,314 minimum inhibitory concentrations matched with genotypes in 92.4% of the cases; however, prediction of phenotype based on genotypic data was only partially efficient, especially with respect to aminoglycosides and chloramphenicol. Our study sheds light on resistance mechanisms and their transferability potential in LAB and bifidobacteria, which will be useful for risk assessment analysis.
© 2023 Rozman et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
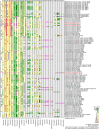

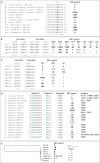



References
-
- World Health Organisation (2011) Tackling Antibiotic Resistance from a Food Safety Perspective in Europe. Copenhagen: WHO Regional Office for Europe.
-
- Zaheer R, Cook SR, Barbieri R, Goji N, Cameron A, Petkau A, Polo RO, Tymensen L, Stamm C, Song J, et al. (2020) Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci Rep 10: 3937. 10.1038/s41598-020-61002-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
