Utilization of n-alkane and roles of lipid transfer proteins in Yarrowia lipolytica
- PMID: 36781616
- PMCID: PMC9925530
- DOI: 10.1007/s11274-023-03541-3
Utilization of n-alkane and roles of lipid transfer proteins in Yarrowia lipolytica
Abstract
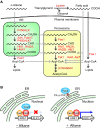
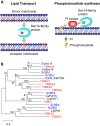
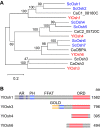
Yarrowia lipolytica, a dimorphic yeast belonging to the Ascomycota, has potent abilities to utilize hydrophobic compounds, such as n-alkanes and fatty acids, as carbon and energy sources. Yarrowia lipolytica can synthesize and accumulate large amounts of lipids, making it a promising host to produce various lipids and convert n-alkanes to useful compounds. For advanced use of Y. lipolytica in these applications, it is necessary to understand the metabolism of these hydrophobic compounds in this yeast and the underlying molecular mechanisms. In this review, current knowledge on the n-alkane metabolism and how this is regulated in Y. lipolytica is summarized. Furthermore, recent studies revealed that lipid transfer proteins are involved in the utilization of n-alkanes and the regulation of cell morphology in response to n-alkanes. This review discusses the roles of membrane lipids in these processes in Y. lipolytica.
Keywords: Dimorphism; Fatty acid; Lipid transfer protein; Yarrowia lipolytica; n-Alkane.
© 2023. The Author(s).
Conflict of interest statement
The author has no conflicts of interest to declare.
Figures
References
-
- Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Non-conventional yeast in biotechnology a handbook. Berlin, Heidelberg, New York: Springer; 1996. pp. 313–388.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

