Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment
- PMID: 36781852
- PMCID: PMC9925748
- DOI: 10.1038/s41467-023-36296-4
Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment
Erratum in
-
Author Correction: Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment.Nat Commun. 2023 Jul 3;14(1):3912. doi: 10.1038/s41467-023-39680-2. Nat Commun. 2023. PMID: 37400453 Free PMC article. No abstract available.
Abstract
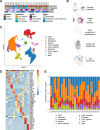
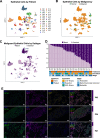
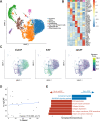
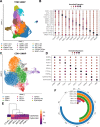
The tumor microenvironment (TME) in pancreatic ductal adenocarcinoma (PDAC) is a complex ecosystem that drives tumor progression; however, in-depth single cell characterization of the PDAC TME and its role in response to therapy is lacking. Here, we perform single-cell RNA sequencing on freshly collected human PDAC samples either before or after chemotherapy. Overall, we find a heterogeneous mixture of basal and classical cancer cell subtypes, along with distinct cancer-associated fibroblast and macrophage subpopulations. Strikingly, classical and basal-like cancer cells exhibit similar transcriptional responses to chemotherapy and do not demonstrate a shift towards a basal-like transcriptional program among treated samples. We observe decreased ligand-receptor interactions in treated samples, particularly between TIGIT on CD8 + T cells and its receptor on cancer cells, and identify TIGIT as the major inhibitory checkpoint molecule of CD8 + T cells. Our results suggest that chemotherapy profoundly impacts the PDAC TME and may promote resistance to immunotherapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
New target and timing in PDAC immunotherapy?Nat Rev Gastroenterol Hepatol. 2023 Apr;20(4):201. doi: 10.1038/s41575-023-00763-6. Nat Rev Gastroenterol Hepatol. 2023. PMID: 36882557 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

