TMEM164 is an acyltransferase that forms ferroptotic C20:4 ether phospholipids
- PMID: 36782012
- PMCID: PMC10362496
- DOI: 10.1038/s41589-022-01253-7
TMEM164 is an acyltransferase that forms ferroptotic C20:4 ether phospholipids
Abstract
Ferroptosis is an iron-dependent form of cell death driven by oxidation of polyunsaturated fatty acid (PUFA) phospholipids. Large-scale genetic screens have uncovered a specialized role for PUFA ether phospholipids (ePLs) in promoting ferroptosis. Understanding of the enzymes involved in PUFA-ePL production, however, remains incomplete. Here we show, using a combination of pathway mining of genetic dependency maps, AlphaFold-guided structure predictions and targeted lipidomics, that the uncharacterized transmembrane protein TMEM164-the genetic ablation of which has been shown to protect cells from ferroptosis-is a cysteine active center enzyme that selectively transfers C20:4 acyl chains from phosphatidylcholine to lyso-ePLs to produce PUFA ePLs. Genetic deletion of TMEM164 across a set of ferroptosis-sensitive cancer cell lines caused selective reductions in C20:4 ePLs with minimal effects on C20:4 diacyl PLs, and this lipid profile produced a variable range of protection from ferroptosis, supportive of an important but contextualized role for C20:4 ePLs in this form of cell death.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
The authors declare no competing financial interests.
Figures


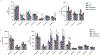
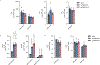




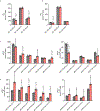





References
-
- van Meer G, Voelker DR & Feigenson GW Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9, 112–124 (2008). https://doi.org: 10.1038/nrm2330 - DOI - PMC - PubMed
-
- Yang WS et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci U S A 113, E4966–4975 (2016). https://doi.org: 10.1073/pnas.1603244113 - DOI - PMC - PubMed
-
- Stockwell BR et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 171, 273–285 (2017). https://doi.org: 10.1016/j.cell.2017.09.021 - DOI - PMC - PubMed
-
- Yang WS et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). https://doi.org: 10.1016/j.cell.2013.12.010 - DOI - PMC - PubMed
-
- Eaton JK et al. Selective covalent targeting of GPX4 using masked nitrile-oxide electrophiles. Nat Chem Biol 16, 497–506 (2020). https://doi.org: 10.1038/s41589-020-0501-5 - DOI - PMC - PubMed
Methods-only References
-
- Cajka T, Smilowitz JT & Fiehn O Validating Quantitative Untargeted Lipidomics Across Nine Liquid Chromatography-High-Resolution Mass Spectrometry Platforms. Anal Chem 89, 12360–12368 (2017). https://doi.org: 10.1021/acs.analchem.7b03404 - DOI - PubMed
-
- Koch J et al. Unequivocal Mapping of Molecular Ether Lipid Species by LC-MS/MS in Plasmalogen-Deficient Mice. Anal Chem 92, 11268–11276 (2020). https://doi.org: 10.1021/acs.analchem.0c01933 - DOI - PMC - PubMed
-
- Huang TP, Newby GA & Liu DR Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat Protoc 16, 1089–1128 (2021). https://doi.org: 10.1038/s41596-020-00450-9 - DOI - PubMed
-
- Clement K et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226 (2019). https://doi.org: 10.1038/s41587-019-0032-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

