Identification of m7G regulator-mediated RNA methylation modification patterns and related immune microenvironment regulation characteristics in heart failure
- PMID: 36782329
- PMCID: PMC9926673
- DOI: 10.1186/s13148-023-01439-3
Identification of m7G regulator-mediated RNA methylation modification patterns and related immune microenvironment regulation characteristics in heart failure
Abstract
Background: N7-methylguanosine (m7G) modification has been reported to regulate RNA expression in multiple pathophysiological processes. However, little is known about its role and association with immune microenvironment in heart failure (HF).
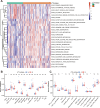
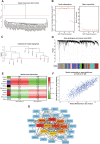
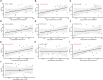
Results: One hundred twenty-four HF patients and 135 nonfailing donors (NFDs) from six microarray datasets in the gene expression omnibus (GEO) database were included to evaluate the expression profiles of m7G regulators. Results revealed that 14 m7G regulators were differentially expressed in heart tissues from HF patients and NFDs. Furthermore, a five-gene m7G regulator diagnostic signature, NUDT16, NUDT4, CYFIP1, LARP1, and DCP2, which can easily distinguish HF patients and NFDs, was established by cross-combination of three machine learning methods, including best subset regression, regularization techniques, and random forest algorithm. The diagnostic value of five-gene m7G regulator signature was further validated in human samples through quantitative reverse-transcription polymerase chain reaction (qRT-PCR). In addition, consensus clustering algorithms were used to categorize HF patients into distinct molecular subtypes. We identified two distinct m7G subtypes of HF with unique m7G modification pattern, functional enrichment, and immune characteristics. Additionally, two gene subgroups based on m7G subtype-related genes were further discovered. Single-sample gene-set enrichment analysis (ssGSEA) was utilized to assess the alterations of immune microenvironment. Finally, utilizing protein-protein interaction network and weighted gene co-expression network analysis (WGCNA), we identified UQCRC1, NDUFB6, and NDUFA13 as m7G methylation-associated hub genes with significant clinical relevance to cardiac functions.
Conclusions: Our study discovered for the first time that m7G RNA modification and immune microenvironment are closely correlated in HF development. A five-gene m7G regulator diagnostic signature for HF (NUDT16, NUDT4, CYFIP1, LARP1, and DCP2) and three m7G methylation-associated hub genes (UQCRC1, NDUFB6, and NDUFA13) were identified, providing new insights into the underlying mechanisms and effective treatments of HF.
Keywords: Bioinformatic analysis; Heart failure; Immune infiltration; Machine learning; N7-methylguanosine; Unsupervised clustering.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

