Long Noncoding RNA PCGEM1 Facilitates Tumor Growth and Metastasis of Osteosarcoma by Sponging miR-433-3p and Targeting OMA1
- PMID: 36782343
- PMCID: PMC10102293
- DOI: 10.1111/os.13648
Long Noncoding RNA PCGEM1 Facilitates Tumor Growth and Metastasis of Osteosarcoma by Sponging miR-433-3p and Targeting OMA1
Abstract
Objective: Osteosarcoma (OS) is regarded as one of the most common malignant bone tumors, mainly occurring in children and adolescents with high mortality. The dysregulation of lncRNAs is reported to regulate tumor development and be closely related to patient prognosis. Nevertheless, the role of long noncoding RNAs (lncRNAs) prostate-specific transcript 1 (PCGEM1) in OS remains uncharacterized. The current study aimed to explore the role of PCGEM1 in OS.
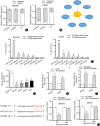
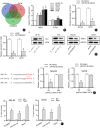
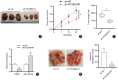
Methods: Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to examine the expression of PCGEM1 in OS cell lines. CCK-8, colony formation, Transwell, and western blotting analyses were applied to measure OS cell viability, proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) after PCGEM1 downregulation. Nuclear-cytoplasmic fractionation, RNA pulldown, RNA immunoprecipitation (RIP), luciferase reporter assays were performed to verify the relationship among PCGEM1, miR-433-3p. and OMA1 in OS. The xenograft tumor models were established to evaluate the effect of PCGEM1 on tumor growth of OS.
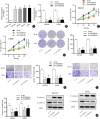
Results: In this study, we discovered that PCGEM1 knockdown inhibited cell proliferation, migration, invasion and EMT in OS (P < 0.05). Additionally, PCGEM1 directly bound to miR-433-3p (P < 0.01). OMA1 was confirmed to be a target gene of miR-433-3p (P < 0.05), positively regulated by PCGEM1 but negatively regulated by miR-433-3p. Rescue assays further verified that overexpression of OMA1 reversed the PCGEM1 knockdown-mediated inhibitory effect on the malignant phenotype in OS cells (P < 0.05). Moreover, knockdown of PCGEM1 inhibited tumor growth and metastasis in vivo (P < 0.05).
Conclusions: Overall, PCGEM1 mediated tumor growth and metastasis of OS by sponging miR-433-3p and regulating OMA1, which might provide an innovative strategy for OS diagnosis or treatment.
Keywords: OMA1; Osteosarcoma; PCGEM1; miR-433-3p.
© 2023 The Authors. Orthopaedic Surgery published by Tianjin Hospital and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–5. - PubMed
-
- Winkler K, Bielack S, Bieling P. Osteosarcoma. Curr Opin Oncol. 1990;2(3):486–90. - PubMed
-
- Simpson E, Brown HL. Understanding osteosarcomas. Jaapa. 2018;31(8):15–9. - PubMed
-
- Bielack SS, Kempf‐Bielack B, Delling G, Exner GU, Flege S, Helmke K, et al. Prognostic factors in high‐grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20(3):776–90. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

