Identification of flucloxacillin-modified hepatocellular proteins: implications in flucloxacillin-induced liver injury
- PMID: 36782357
- PMCID: PMC10371196
- DOI: 10.1093/toxsci/kfad015
Identification of flucloxacillin-modified hepatocellular proteins: implications in flucloxacillin-induced liver injury
Abstract
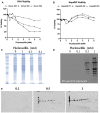
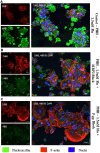
Flucloxacillin is a β-lactam antibiotic associated with a high incidence of drug-induced liver injury. Although expression of HLA-B*57:01 is associated with increased susceptibility, little is known of the pathological mechanisms involved in the induction of the clinical phenotype. Irreversible protein modification is suspected to drive the reaction through the provision of flucloxacillin-modified peptides that are presented to T-cells by the protein encoded by the risk allele. In this study, we have shown that flucloxacillin binds to multiple proteins within human primary hepatocytes, including major hepatocellular proteins (hemoglobin and albumin) and mitochondrial proteins. Inhibition of membrane transporters multidrug resistance-associated protein 2 (MRP2) and P-glycoprotein (P-gp) appeared to reduce the levels of covalent binding. A diverse range of proteins with different functions was found to be targeted by flucloxacillin, including adaptor proteins (14-3-3), proteins with catalytic activities (liver carboxylesterase 1, tRNA-splicing endonuclease subunit Sen2, All-trans-retinol dehydrogenase ADH1B, Glutamate dehydrogenase 1 mitochondrial, Carbamoyl-phosphate synthase [ammonia] mitochondrial), and transporters (hemoglobin, albumin, and UTP-glucose-1-phosphate uridylyltransferase). These flucloxacillin-modified intracellular proteins could provide a potential source of neoantigens for HLA-B*57:01 presentation by hepatocytes. More importantly, covalent binding to critical cellular proteins could be the molecular initiating events that lead to flucloxacillin-induced cholestasis Data are available via ProteomeXchange with identifier PXD038581.
Keywords: covalent binding; flucloxacillin DILI; hepatocytes; membrane transporters.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Society of Toxicology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Baldo B. A., Pham N. H., Weiner J. (1995). Detection and side-chain specificity of IgE antibodies to flucloxacillin in allergic subjects. J. Mol. Recognit. 8, 171–177. - PubMed
-
- Daly A. K., Donaldson P. T., Bhatnagar P., Shen Y., Pe'er I., Floratos A., Daly M. J., Goldstein D. B., John S., Nelson M. R., for the DILIGEN study., et al. (2009). HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 41, 816–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

