Production and Characterization of a SARS-CoV-2 Nucleocapsid Protein Reference Material
- PMID: 36785774
- PMCID: PMC9662649
- DOI: 10.1021/acsmeasuresciau.2c00050
Production and Characterization of a SARS-CoV-2 Nucleocapsid Protein Reference Material
Abstract
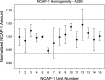
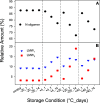
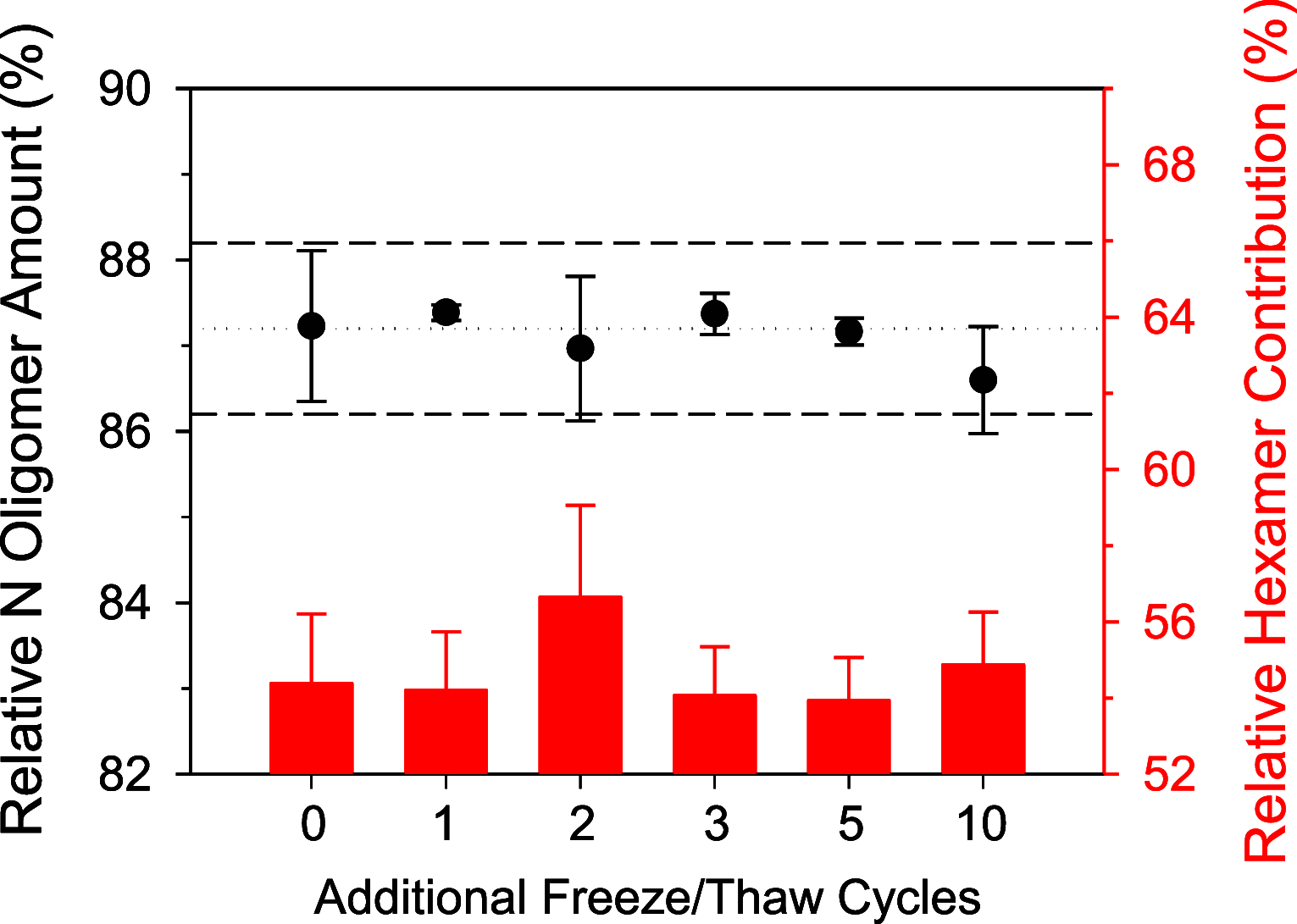
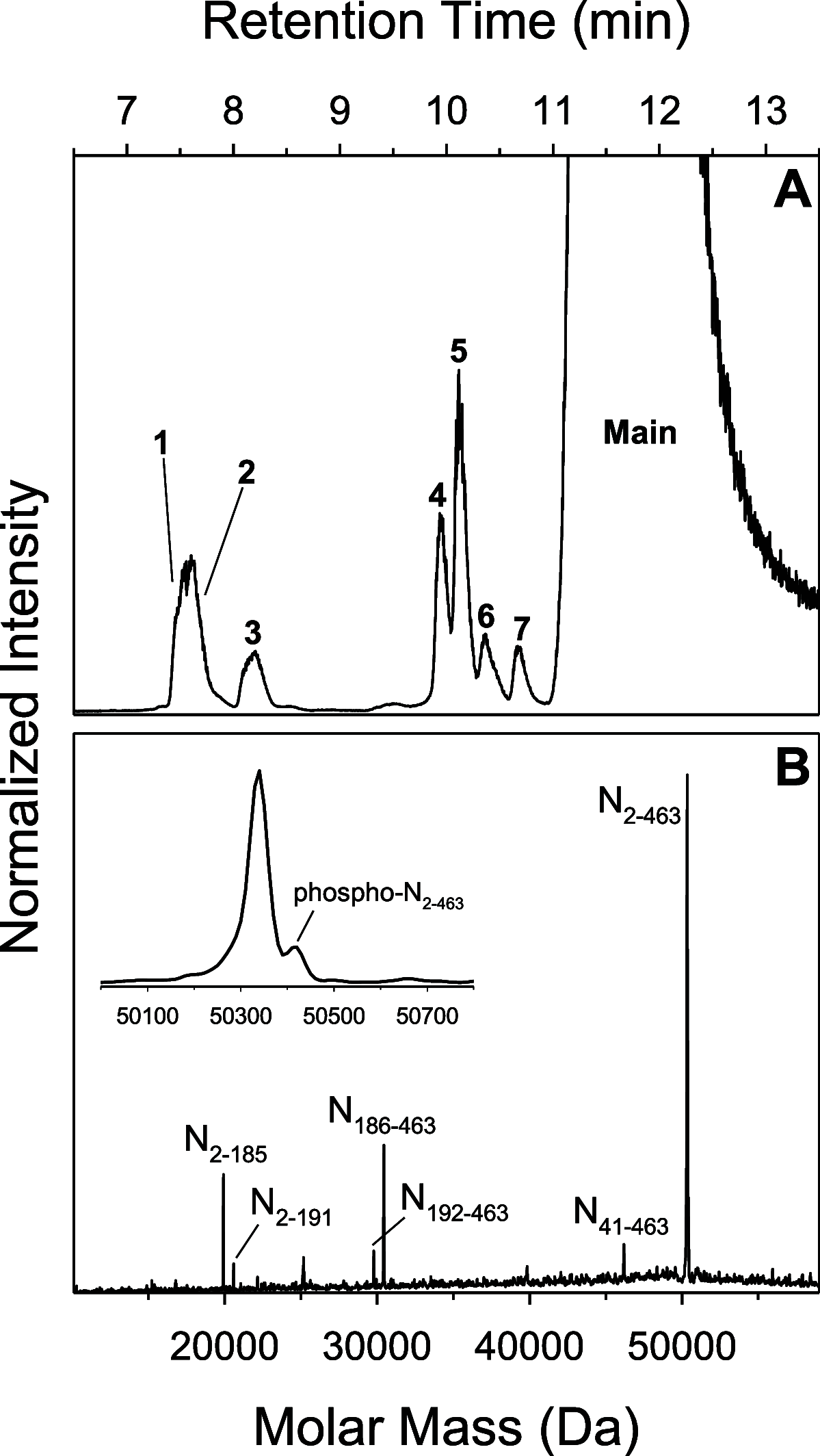
Rapid antigen tests have become a widely used COVID-19 diagnostic tool with demand accelerating in response to the highly contagious SARS-CoV-2 Omicron variant. Hundreds of such test kits are approved for use worldwide, predominantly reporting on the presence of the viral nucleocapsid (N) protein, yet the comparability among manufacturers remains unclear and the need for reference standards is recognized. To address this lack of standardization, the National Research Council Canada has developed a SARS-CoV-2 nucleocapsid protein reference material solution, NCAP-1. Reference value determination for N protein content was realized by amino acid analysis (AAA) via double isotope dilution liquid chromatography-tandem mass spectrometry (LC-ID-MS/MS) following acid hydrolysis of the protein, in conjunction with UV spectrophotometry based on tryptophan and tyrosine absorbance at 280 nm. The homogeneity of the material was established through spectrophotometric absorbance readings at 280 nm. The molar concentration of the N protein in NCAP-1 was 10.0 ± 1.9 μmol L-1 (k = 2, 95% confidence interval). Reference mass concentration and mass fraction values were subsequently calculated using the protein molecular weight and density of the NCAP-1 solution. Changes to protein higher-order structure, probed by size-exclusion liquid chromatography (LC-SEC) with UV detection, were used to evaluate transportation and storage stabilities. LC-SEC revealed nearly 90% of the N protein in the material is present as a mixture of hexamers and tetramers. The remaining low molecular weight species (<30 kDa) were interrogated by top-down mass spectrometry and determined to be autolysis products homologous to those previously documented for N protein of the original SARS-CoV [Biochem. Biophys. Res. Commun.2008t, 377, 429-433].
Crown © 2022. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Johns Hopkins Coronavirus Resource Center. https://coronavirus.jhu.edu (accessed 24 June 2022).
-
- Chi X.; Yan R.; Zhang J.; Zhang G.; Zhang Y.; Hao M.; Zhang Z.; Fan P.; Dong Y.; Yang Y.; Chen Z.; Guo Y.; Zhang J.; Li Y.; Song X.; Chen Y.; Xia L.; Fu L.; Hou L.; Xu J.; Yu C.; Li J.; Zhou Q.; Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. 10.1126/science.abc6952. - DOI - PMC - PubMed
-
- Hansen J.; Baum A.; Pascal K. E.; Russo V.; Giordano S.; Elzbieta Wloga E.; Fulton B. O.; Yan Y.; Koon K.; Patel K.; Chung K. M.; Hermann A.; Ullman E.; Cruz J.; Rafique A.; Huang T.; Fairhurst J.; Libertiny C.; Malbec M.; Lee W.-Y.; Welsh R.; Farr G.; Pennington S.; Deshpande D.; Cheng J.; Watty A.; Bouffard P.; Babb R.; Levenkova N.; Chen C.; Zhang B.; Romero Hernandez A.; Saotome K.; Zhou Y.; Franklin M.; Sivapalasingam S.; Chien Lye D.; Weston S.; Logue J.; Haupt R.; Frieman M.; Chen G.; Olson W.; Murphy A. J.; Stahl N.; Yancopoulos G. D.; Kyratsous C. A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, 369, 1010–1014. 10.1126/science.abd0827. - DOI - PMC - PubMed
-
- Cao Y.; Wang J.; Jian F.; Xiao T.; Song W.; Yisimayi A.; Huang W.; Li Q.; Wang P.; An R.; Wang J.; Niu X.; Yang S.; Liang H.; Sun H.; Li T.; Yu Y.; Cui Q.; Liu S.; Yang X.; Du S.; Zhang Z.; Hao X.; Shao F.; Jin R.; Wang X.; Xiao J.; Wang Y.; Sunney Xie X. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. 10.1038/s41586-021-04385-3. - DOI - PMC - PubMed
-
- FDA Emergency Use Authorizations. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-em... (accessed 21 February 2022).
LinkOut - more resources
Full Text Sources
Miscellaneous
