Biotechnologically potential genes in a polysaccharide-degrading epibiont of the Indonesian brown algae Hydroclathrus sp
- PMID: 36786886
- PMCID: PMC9928984
- DOI: 10.1186/s43141-023-00461-5
Biotechnologically potential genes in a polysaccharide-degrading epibiont of the Indonesian brown algae Hydroclathrus sp
Abstract
Background: Marine bacteria have recently attracted increasing attention to be harnessed for the production of valuable enzymes, vitamins, and bioactive compounds. Bacteria associated with the surfaces of marine macroalgae, called epibionts, are particularly interesting from ecological and biotechnological points of view, as they often exhibit antimicrobial activities to compete with pathogenic bacteria for nutrients and spaces. In search for biotechnologically potential genes from marine bacteria, we sequenced and analysed the genome of the epibiont HI03-3b, a polysaccharide-degrading bacterium associated with the surface of the Indonesian brown algae Hydroclathrus sp.
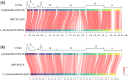
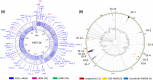
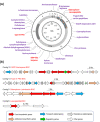
Results: The algal epibiont HI03-3b has a genome of approximately 4,860,704 bp in size with 42.02 mol% G + C content, consisting of 5655 open reading frames (ORFs), 4409 genes coding for proteins (CDSs), 94 genes for tRNAs, and 32 genes for rRNAs. The genome sequence of HI03-3b was most closely related to that of Cytobacillus firmus NCTC10335 with the average amino acid identity (AAI) of 95.0 %, average nucleotide identity (ANI) of 94.1 %, and a recommended DNA-DNA hybridization (DDH) of 57.60 %. These scores are lower than the most frequently used standard for species demarcation (95% ANI cutoff) and the new species threshold (DDH > 70.0% for the same bacterial species). Some differences in genome features and gene composition were observed between HI03-3b and NCTC10335, such as genes encoding carbohydrate active enzymes. These suggest that HI03-3b is unique and likely a novel species within Cytobacillus genus, and we therefore proposed its name as Cytobacillus wakatobiense HI03-3b. Genome sequence analyses indicated the presence of genes involved not only in polysaccharide and protein degradation but also in vitamin and secondary metabolite biosynthesis. Some of them encode enzymes and compounds with biotechnological interest, such as protease, chitinase, subtilisin, pullulanase, and bacillolysin, which are often associated with antimicrobial or antibiofilm activities. This antimicrobial potential is supported by our finding that the extracellular protein fraction of this epibiont inhibited the growth of the bacterial pathogen Staphylococcus aureus.
Conclusion: The epibiont Cytobacillus HI03-3b harbours genes for polysaccharide and protein degradation as well as for natural product biosynthesis, suggesting its potential ecological roles in outcompeting other bacteria during biofilm formation as well as in protecting its algal host from predation. Due to the presence of genes for vitamin biosynthesis, it might also provide the algal host with vitamins for growth and development. Some of these metabolic genes are biotechnologically important, as they could become a platform for bioengineering to generate various seaweed-derived substances sustainably, such as antibiofilm agents and vitamins, which are beneficial for human health.
Keywords: Antimicrobial activity; Bacterial epibiont; Biotechnologically potential genes; Ecological role; Genome sequencing; Marine macroalgae.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
