FinO/ProQ-family proteins: an evolutionary perspective
- PMID: 36787218
- PMCID: PMC9977716
- DOI: 10.1042/BSR20220313
FinO/ProQ-family proteins: an evolutionary perspective
Abstract
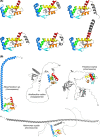
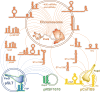
RNA-binding proteins are key actors of post-transcriptional networks. Almost exclusively studied in the light of their interactions with RNA ligands and the associated functional events, they are still poorly understood as evolutionary units. In this review, we discuss the FinO/ProQ family of bacterial RNA chaperones, how they evolve and spread across bacterial populations and what properties and opportunities they provide to their host cells. We reflect on major conserved and divergent themes within the family, trying to understand how the same ancestral RNA-binding fold, augmented with additional structural elements, could yield either highly specialised proteins or, on the contrary, globally acting regulatory hubs with a pervasive impact on gene expression. We also consider dominant convergent evolutionary trends that shaped their RNA chaperone activity and recurrently implicated the FinO/ProQ-like proteins in bacterial DNA metabolism, translation and virulence. Finally, we offer a new perspective in which FinO/ProQ-family regulators emerge as active evolutionary players with both negative and positive roles, significantly impacting the evolutionary modes and trajectories of their bacterial hosts.
Keywords: FinO; ProQ; RNA chaperone; RNA-binding proteins; evolution.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures




Similar articles
-
Different RNA recognition by ProQ and FinO depends on the sequence surrounding intrinsic terminator hairpins.RNA. 2025 Apr 16;31(5):692-708. doi: 10.1261/rna.080206.124. RNA. 2025. PMID: 40044219 Free PMC article.
-
ProQ is an RNA chaperone that controls ProP levels in Escherichia coli.Biochemistry. 2011 Apr 19;50(15):3095-106. doi: 10.1021/bi101683a. Epub 2011 Mar 23. Biochemistry. 2011. PMID: 21381725
-
ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers?Mol Microbiol. 2017 Jun;104(6):905-915. doi: 10.1111/mmi.13679. Epub 2017 May 3. Mol Microbiol. 2017. PMID: 28370625 Free PMC article. Review.
-
Solution structure and RNA-binding of a minimal ProQ-homolog from Legionella pneumophila (Lpp1663).RNA. 2020 Dec;26(12):2031-2043. doi: 10.1261/rna.077354.120. Epub 2020 Sep 28. RNA. 2020. PMID: 32989045 Free PMC article.
-
The FinO family of bacterial RNA chaperones.Plasmid. 2015 Mar;78:79-87. doi: 10.1016/j.plasmid.2014.07.003. Epub 2014 Aug 4. Plasmid. 2015. PMID: 25102058 Review.
Cited by
-
The Early Evolution of Tudor Genes in Holozoa and How Their Distribution Was Influenced by Life History Traits in Metazoa.Genome Biol Evol. 2025 May 30;17(6):evaf051. doi: 10.1093/gbe/evaf051. Genome Biol Evol. 2025. PMID: 40489295 Free PMC article.
-
ProQ prevents mRNA degradation through inhibition of poly(A) polymerase.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf103. doi: 10.1093/nar/gkaf103. Nucleic Acids Res. 2025. PMID: 40036335 Free PMC article.
-
Antimicrobial resistance and mechanisms of epigenetic regulation.Front Cell Infect Microbiol. 2023 Jun 14;13:1199646. doi: 10.3389/fcimb.2023.1199646. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37389209 Free PMC article. Review.
-
Generation of single-cysteine E. coli ProQ variants to study RNA-protein interaction mechanisms.MicroPubl Biol. 2024 Apr 9;2024:10.17912/micropub.biology.001188. doi: 10.17912/micropub.biology.001188. eCollection 2024. MicroPubl Biol. 2024. PMID: 38660567 Free PMC article.
-
The FinO/ProQ-like protein PA2582 impacts antimicrobial resistance in Pseudomonas aeruginosa.Front Microbiol. 2024 Jun 26;15:1422742. doi: 10.3389/fmicb.2024.1422742. eCollection 2024. Front Microbiol. 2024. PMID: 39011145 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

