Split aminoacyl-tRNA synthetases for proximity-induced stop codon suppression
- PMID: 36787361
- PMCID: PMC9974479
- DOI: 10.1073/pnas.2219758120
Split aminoacyl-tRNA synthetases for proximity-induced stop codon suppression
Abstract
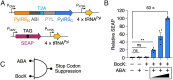
Synthetic biology tools for regulating gene expression have many useful biotechnology and therapeutic applications. Most tools developed for this purpose control gene expression at the level of transcription, and relatively few methods are available for regulating gene expression at the translational level. Here, we design and engineer split orthogonal aminoacyl-tRNA synthetases (o-aaRS) as unique tools to control gene translation in bacteria and mammalian cells. Using chemically induced dimerization domains, we developed split o-aaRSs that mediate gene expression by conditionally suppressing stop codons in the presence of the small molecules rapamycin and abscisic acid. By activating o-aaRSs, these molecular switches induce stop codon suppression, and in their absence stop codon suppression is turned off. We demonstrate, in Escherichia coli and in human cells, that split o-aaRSs function as genetically encoded AND gates where stop codon suppression is controlled by two distinct molecular inputs. In addition, we show that split o-aaRSs can be used as versatile biosensors to detect therapeutically relevant protein-protein interactions, including those involved in cancer, and those that mediate severe acute respiratory syndrome-coronavirus-2 infection.
Keywords: genetic code expansion; noncanonical amino acids; pyrrolysyl-tRNA synthetase; stop codon suppression; synthetic biology.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Kent R., Dixon N., Contemporary tools for regulating gene expression in bacteria. Trends Biotechnol. 38, 316–333 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

