A fair experimental comparison of neural network architectures for latent representations of multi-omics for drug response prediction
- PMID: 36788531
- PMCID: PMC9926634
- DOI: 10.1186/s12859-023-05166-7
A fair experimental comparison of neural network architectures for latent representations of multi-omics for drug response prediction
Abstract
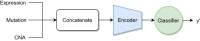
Background: Recent years have seen a surge of novel neural network architectures for the integration of multi-omics data for prediction. Most of the architectures include either encoders alone or encoders and decoders, i.e., autoencoders of various sorts, to transform multi-omics data into latent representations. One important parameter is the depth of integration: the point at which the latent representations are computed or merged, which can be either early, intermediate, or late. The literature on integration methods is growing steadily, however, close to nothing is known about the relative performance of these methods under fair experimental conditions and under consideration of different use cases.
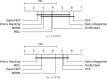
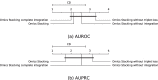
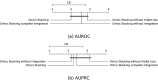
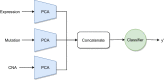
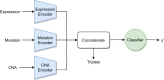
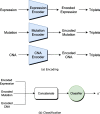
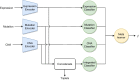
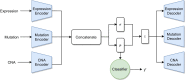
Results: We developed a comparison framework that trains and optimizes multi-omics integration methods under equal conditions. We incorporated early integration, PCA and four recently published deep learning methods: MOLI, Super.FELT, OmiEmbed, and MOMA. Further, we devised a novel method, Omics Stacking, that combines the advantages of intermediate and late integration. Experiments were conducted on a public drug response data set with multiple omics data (somatic point mutations, somatic copy number profiles and gene expression profiles) that was obtained from cell lines, patient-derived xenografts, and patient samples. Our experiments confirmed that early integration has the lowest predictive performance. Overall, architectures that integrate triplet loss achieved the best results. Statistical differences can, overall, rarely be observed, however, in terms of the average ranks of methods, Super.FELT is consistently performing best in a cross-validation setting and Omics Stacking best in an external test set setting.
Conclusions: We recommend researchers to follow fair comparison protocols, as suggested in the paper. When faced with a new data set, Super.FELT is a good option in the cross-validation setting as well as Omics Stacking in the external test set setting. Statistical significances are hardly observable, despite trends in the algorithms' rankings. Future work on refined methods for transfer learning tailored for this domain may improve the situation for external test sets. The source code of all experiments is available under https://github.com/kramerlab/Multi-Omics_analysis.
Keywords: Autoencoder; Deep learning; Drug response; Machine learning; Multi-omics; Neural network.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Super.FELT: supervised feature extraction learning using triplet loss for drug response prediction with multi-omics data.BMC Bioinformatics. 2021 May 25;22(1):269. doi: 10.1186/s12859-021-04146-z. BMC Bioinformatics. 2021. PMID: 34034645 Free PMC article.
-
MOLI: multi-omics late integration with deep neural networks for drug response prediction.Bioinformatics. 2019 Jul 15;35(14):i501-i509. doi: 10.1093/bioinformatics/btz318. Bioinformatics. 2019. PMID: 31510700 Free PMC article.
-
moBRCA-net: a breast cancer subtype classification framework based on multi-omics attention neural networks.BMC Bioinformatics. 2023 Apr 26;24(1):169. doi: 10.1186/s12859-023-05273-5. BMC Bioinformatics. 2023. PMID: 37101124 Free PMC article.
-
Deep learning-based approaches for multi-omics data integration and analysis.BioData Min. 2024 Oct 2;17(1):38. doi: 10.1186/s13040-024-00391-z. BioData Min. 2024. PMID: 39358793 Free PMC article. Review.
-
Data Integration Using Advances in Machine Learning in Drug Discovery and Molecular Biology.Methods Mol Biol. 2021;2190:167-184. doi: 10.1007/978-1-0716-0826-5_7. Methods Mol Biol. 2021. PMID: 32804365 Review.
Cited by
-
Visible neural networks for multi-omics integration: a critical review.Front Artif Intell. 2025 Jul 17;8:1595291. doi: 10.3389/frai.2025.1595291. eCollection 2025. Front Artif Intell. 2025. PMID: 40746431 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

