Centriolar satellites expedite mother centriole remodeling to promote ciliogenesis
- PMID: 36790165
- PMCID: PMC9998092
- DOI: 10.7554/eLife.79299
Centriolar satellites expedite mother centriole remodeling to promote ciliogenesis
Abstract
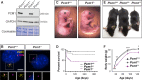
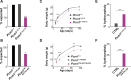
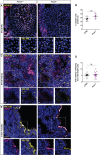
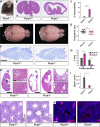
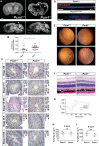
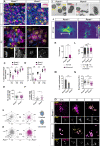
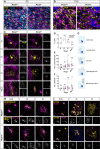
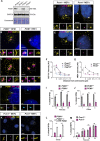
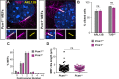
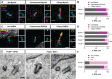
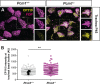
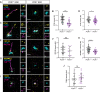
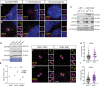
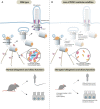
Centrosomes are orbited by centriolar satellites, dynamic multiprotein assemblies nucleated by Pericentriolar material 1 (PCM1). To study the requirement for centriolar satellites, we generated mice lacking PCM1, a crucial component of satellites. Pcm1-/- mice display partially penetrant perinatal lethality with survivors exhibiting hydrocephalus, oligospermia, and cerebellar hypoplasia, and variably expressive phenotypes such as hydronephrosis. As many of these phenotypes have been observed in human ciliopathies and satellites are implicated in cilia biology, we investigated whether cilia were affected. PCM1 was dispensable for ciliogenesis in many cell types, whereas Pcm1-/- multiciliated ependymal cells and human PCM1-/- retinal pigmented epithelial 1 (RPE1) cells showed reduced ciliogenesis. PCM1-/- RPE1 cells displayed reduced docking of the mother centriole to the ciliary vesicle and removal of CP110 and CEP97 from the distal mother centriole, indicating compromised early ciliogenesis. Similarly, Pcm1-/- ependymal cells exhibited reduced removal of CP110 from basal bodies in vivo. We propose that PCM1 and centriolar satellites facilitate efficient trafficking of proteins to and from centrioles, including the departure of CP110 and CEP97 to initiate ciliogenesis, and that the threshold to trigger ciliogenesis differs between cell types.
Keywords: centriolar satellites; centriole; centrosome; cilia; ciliary vesicle; ciliogenesis; condensates; developmental biology; human; mouse.
© 2023, Hall, Kumar et al.
Conflict of interest statement
EH, DK, SP, PY, VH, JG, LR, LM, DD, PT, RM, LM, MF, GG, LW, TQ, LP, PM No competing interests declared, JR Reviewing editor, eLife
Figures









Update of
References
-
- Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW. QuPath: open source software for digital pathology image analysis. Scientific Reports. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM095941/GM/NIGMS NIH HHS/United States
- R01AR054396/NH/NIH HHS/United States
- 5K99GM140175/NH/NIH HHS/United States
- R01 HD089918/HD/NICHD NIH HHS/United States
- R01GM095941/NH/NIH HHS/United States
- 702601/MCCC_/Marie Curie/United Kingdom
- RO1HD089918/NH/NIH HHS/United States
- R01 DE029454/DE/NIDCR NIH HHS/United States
- MR_UU_1201018/26/MRC_/Medical Research Council/United Kingdom
- MC_UU_12018/26/MRC_/Medical Research Council/United Kingdom
- K99 GM140175/GM/NIGMS NIH HHS/United States
- MC_UU_00007/14/MRC_/Medical Research Council/United Kingdom
- R01 AR054396/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

