Germline variation in RASAL2 may predict survival in patients with RAS-activated colorectal cancer
- PMID: 36790221
- PMCID: PMC10805173
- DOI: 10.1002/gcc.23133
Germline variation in RASAL2 may predict survival in patients with RAS-activated colorectal cancer
Abstract
Background: Therapeutic agents that specifically target patients with RAS mutant colorectal cancer (CRC) are needed. We sought potential drug targets by relating genome-wide association study and survival data in patients with advanced CRC profiled for mitogen-activated protein kinase (MAPK) pathway mutations.
Methods: In total, 694 patients from the clinical trials COIN and COIN-B had MAPK-activated CRCs (assigned as KRAS, NRAS, or BRAF mutant). Genome-wide single nucleotide polymorphism (SNP), gene, and gene-set analyses were performed to identify determinants of survival. For rs12028023 in RAS protein activator-like 2 (RASAL2), we studied its effect by MAPK pathway activation status (by comparing to 760 patients without MAPK-activated CRCs), MAPK gene mutation status, surface area of the primary tumor (as a marker of proliferation), and expression on RASAL2.
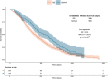
Results: In MAGMA genome-wide analyses, RASAL2 was the most significant gene associated with survival (p = 2.0 × 10-5 ). Patients carrying the minor (A) allele in the lead SNP, rs12028023 in intron 1 of RASAL2, had a median increase in survival of 167 days as compared with patients carrying the major allele. rs12028023 was predictive for survival by MAPK-activation status (pZ-test = 2.1 × 10-3 ). Furthermore, rs12028023 improved survival in patients with RAS mutant (hazard ratio [HR] = 0.62, 95% confidence intervals [CI] = 0.5-0.8, p = 3.4 × 10-5 ) but not BRAF mutant (p = 0.87) CRCs. The rs12028023 A-allele was associated with reduced surface area of the primary tumor (Beta = -0.037, standard error [SE] = 0.017, p = 3.2 × 10-2 ) and reduced RASAL2 expression in cultured fibroblasts (p = 1.6 × 10-11 ).
Conclusion: Our data demonstrate a prognostic role for RASAL2 in patients with MAPK-activated CRCs, with potential as a therapeutic target.
Keywords: MAPK-activation; RAS; RASAL2; colorectal cancer; survival.
© 2023 The Authors. Genes, Chromosomes and Cancer published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Stintzing S, Modest DP, Rossius L, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE‐3): a post‐hoc analysis of tumour dynamics in the final RAS wild‐type subgroup of this randomised open‐label phase 3 trial. Lancet Oncol. 2016;17(10):1426‐1434. doi:10.1016/s1470-2045(16)30269-8 - DOI - PubMed
-
- Khattak MA, Martin H, Davidson A, Phillips M. Role of first‐line anti‐epidermal growth factor receptor therapy compared with anti‐vascular endothelial growth factor therapy in advanced colorectal cancer: a meta‐analysis of randomized clinical trials. Clin Colorectal Cancer. 2015;14(2):81‐90. doi:10.1016/j.clcc.2014.12.011 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

