Intrathymic AAV delivery results in therapeutic site-specific integration at TCR loci in mice
- PMID: 36790505
- PMCID: PMC10356579
- DOI: 10.1182/blood.2022017378
Intrathymic AAV delivery results in therapeutic site-specific integration at TCR loci in mice
Abstract
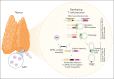
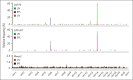
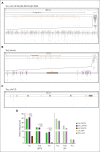
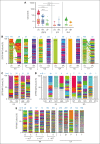
Adeno-associated virus (AAV) vectors have been successfully exploited in gene therapy applications for the treatment of several genetic disorders. AAV is considered an episomal vector, but it has been shown to integrate within the host cell genome after the generation of double-strand DNA breaks or nicks. Although AAV integration raises some safety concerns, it can also provide therapeutic benefit; the direct intrathymic injection of an AAV harboring a therapeutic transgene results in integration in T-cell progenitors and long-term T-cell immunity. To assess the mechanisms of AAV integration, we retrieved and analyzed hundreds of AAV integration sites from lymph node-derived mature T cells and compared these with liver and brain tissue from treated mice. Notably, we found that although AAV integrations in the liver and brain were distributed across the entire mouse genome, >90% of the integrations in T cells were clustered within the T-cell receptor α, β, and γ genes. More precisely, the insertion mapped to DNA breaks created by the enzymatic activity of recombination activating genes (RAGs) during variable, diversity, and joining recombination. Our data indicate that RAG activity during T-cell receptor maturation induces a site-specific integration of AAV genomes and opens new therapeutic avenues for achieving long-term AAV-mediated gene transfer in dividing cells.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



Comment in
-
Intrathymic AAV gene delivery.Blood. 2023 May 11;141(19):2286-2288. doi: 10.1182/blood.2023019863. Blood. 2023. PMID: 37166928 No abstract available.
References
-
- Fisher KJ, Jooss K, Alston J, et al. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3(3):306–312. - PubMed
-
- Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34(3):297–302. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

