Ribosome dysfunction underlies SLFN14-related thrombocytopenia
- PMID: 36790527
- PMCID: PMC10646786
- DOI: 10.1182/blood.2022017712
Ribosome dysfunction underlies SLFN14-related thrombocytopenia
Abstract
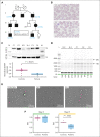
Pathogenic missense variants in SLFN14, which encode an RNA endoribonuclease protein that regulates ribosomal RNA (rRNA) degradation, are known to cause inherited thrombocytopenia (TP) with impaired platelet aggregation and adenosine triphosphate secretion. Despite mild laboratory defects, the patients displayed an obvious bleeding phenotype. However, the function of SLFN14 in megakaryocyte (MK) and platelet biology remains unknown. This study aimed to model the disease in an immortalized MK cell line (imMKCL) and to characterize the platelet transcriptome in patients with the SLFN14 K219N variant. MK derived from heterozygous and homozygous SLFN14 K219N imMKCL and stem cells of blood from patients mainly presented with a defect in proplatelet formation and mitochondrial organization. SLFN14-defective platelets and mature MK showed signs of rRNA degradation; however, this was absent in undifferentiated imMKCL cells and granulocytes. Total platelet RNA was sequenced in 2 patients and 19 healthy controls. Differential gene expression analysis yielded 2999 and 2888 significantly (|log2 fold change| >1, false discovery rate <0.05) up- and downregulated genes, respectively. Remarkably, these downregulated genes were not enriched in any biological pathway, whereas upregulated genes were enriched in pathways involved in (mitochondrial) translation and transcription, with a significant upregulation of 134 ribosomal protein genes (RPGs). The upregulation of mitochondrial RPGs through increased mammalian target of rapamycin complex 1 (mTORC1) signaling in SLFN14 K219N MK seems to be a compensatory response to rRNA degradation. mTORC1 inhibition with rapamycin resulted in further enhanced rRNA degradation in SLFN14 K219N MK. Taken together, our study indicates dysregulation of mTORC1 coordinated ribosomal biogenesis is the disease mechanism for SLFN14-related TP.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures






Comment in
-
SLFN14 ribosomopathy and platelet dysfunction.Blood. 2023 May 4;141(18):2170-2172. doi: 10.1182/blood.2023019949. Blood. 2023. PMID: 37140956 Free PMC article. No abstract available.
References
-
- Liu F, Zhou P, Wang Q, Zhang M, Li D. The Schlafen family: complex roles in different cell types and virus replication. Cell Biol Int. 2018;42(1):2–8. - PubMed
-
- Marconi C, di Buduo CA, Barozzi S, et al. SLFN14-related thrombocytopenia: identification within a large series of patients with inherited thrombocytopenia. Thromb Haemost. 2016;115(5):1076–1079. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

