Genomic analysis reveals geography rather than culture as the predominant factor shaping genetic variation in northern Kenyan human populations
- PMID: 36790743
- PMCID: PMC9949739
- DOI: 10.1002/ajpa.24521
Genomic analysis reveals geography rather than culture as the predominant factor shaping genetic variation in northern Kenyan human populations
Abstract
Objectives: The aim of this study was to characterize the genetic relationships within and among four neighboring ethnolinguistic groups in northern Kenya in light of cultural relationships to understand the extent to which geography and culture shape patterns of genetic variation.
Materials and methods: We collected DNA and demographic information pertaining to aspects of social identity and heritage from 572 individuals across the Turkana, Samburu, Waso Borana, and Rendille of northern Kenya. We sampled individuals across a total of nine clans from these four groups and, additionally, three territorial sections within the Turkana and successfully genotyped 376 individuals.
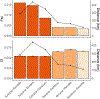
Results: Here we report that geography predominately shapes genetic variation within and among human groups in northern Kenya. We observed a clinal pattern of genetic variation that mirrors the overall geographic distribution of the individuals we sampled. We also found relatively higher rates of intermarriage between the Rendille and Samburu and evidence of gene flow between them that reflect these higher rates of intermarriage. Among the Turkana, we observed strong recent genetic substructuring based on territorial section affiliation. Within ethnolinguistic groups, we found that Y chromosome haplotypes do not consistently cluster by natal clan affiliation. Finally, we found that sampled populations that are geographically closer have lower genetic differentiation, and that cultural similarity does not predict genetic similarity as a whole across these northern Kenyan populations.
Discussion: Overall, the results from this study highlight the importance of geography, even on a local geographic scale, in shaping observed patterns of genetic variation in human populations.
Keywords: Africa; Kenya; cultural FST; culture; genetic FST; genetic structure; geography; social organization.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures


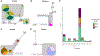


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
