BioID2 screening identifies KIAA1671 as an EPS8 proximal factor that marks sites of microvillus growth
- PMID: 36790915
- PMCID: PMC10092648
- DOI: 10.1091/mbc.E22-11-0498
BioID2 screening identifies KIAA1671 as an EPS8 proximal factor that marks sites of microvillus growth
Abstract
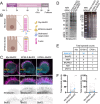
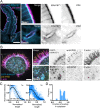
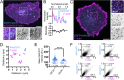
Microvilli are defining morphological features of the apical surfaces in diverse epithelial tissues. To develop our understanding of microvillus biogenesis, we used a biotin proximity-labeling approach to uncover new molecules enriched near EPS8, a well-studied marker of the microvillus distal tip compartment. Mass spectrometry of biotinylated hits identified KIAA1671, a large (∼200 kDa), disordered, and previously uncharacterized protein. Based on immunofluorescent staining and expression of fluorescent protein-tagged constructs, we found that KIAA1671 localizes to the base of the brush border in native intestinal tissue and polarized epithelial-cell culture models, as well as dynamic actin-rich structures in unpolarized, nonepithelial cell types. Live imaging also revealed that during the early stages of microvillar growth, KIAA1671 colocalizes with EPS8 in diffraction-limited puncta. However, once elongation of the core bundle begins, these two factors separate, with EPS8 tracking the distal end and KIAA1671 remaining behind at the base of the structure. These results suggest that KIAA1671 cooperates with EPS8 and potentially other assembly factors to initiate growth of microvilli on the apical surface. These findings offer new details on how transporting epithelial cells builds the brush border and may inform our understanding of how apical specializations are assembled in other epithelial contexts.
Figures


References
-
- Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, Clevers HC (2004). Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116, 457–466. - PubMed
-
- Berryman M, Franck Z, Bretscher A (1993). Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci 105, 1025–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

