ALK fusion NSCLC oncogenes promote survival and inhibit NK cell responses via SERPINB4 expression
- PMID: 36791109
- PMCID: PMC9974509
- DOI: 10.1073/pnas.2216479120
ALK fusion NSCLC oncogenes promote survival and inhibit NK cell responses via SERPINB4 expression
Abstract
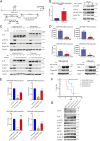
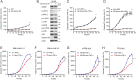
Anaplastic lymphoma kinase (ALK) fusion variants in Non-Small Cell Lung Cancer (NSCLC) consist of numerous dimerizing fusion partners. Retrospective investigations suggest that treatment benefit in response to ALK tyrosine kinase inhibitors (TKIs) differs dependent on the fusion variant present in the patient tumor. Therefore, understanding the oncogenic signaling networks driven by different ALK fusion variants is important. To do this, we developed controlled inducible cell models expressing either Echinoderm Microtubule Associated Protein Like 4 (EML4)-ALK-V1, EML4-ALK-V3, Kinesin Family Member 5B (KIF5B)-ALK, or TRK-fused gene (TFG)-ALK and investigated their transcriptomic and proteomic responses to ALK activity modulation together with patient-derived ALK-positive NSCLC cell lines. This allowed identification of both common and isoform-specific responses downstream of these four ALK fusions. An inflammatory signature that included upregulation of the Serpin B4 serine protease inhibitor was observed in both ALK fusion inducible and patient-derived cells. We show that Signal transducer and activator of transcription 3 (STAT3), Nuclear Factor Kappa B (NF-κB) and Activator protein 1 (AP1) are major transcriptional regulators of SERPINB4 downstream of ALK fusions. Upregulation of SERPINB4 promotes survival and inhibits natural killer cell-mediated cytotoxicity, which has potential for therapeutic impact targeting the immune response together with ALK TKIs in NSCLC.
Keywords: ALK fusion; NGS; anaplastic lymphoma kinase; non-small cell lung cancer; phosphoprofiling.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Rikova K., et al. , Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007). - PubMed
-
- Soda M., et al. , Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007). - PubMed
-
- Zhang S. S., Nagasaka M., Zhu V. W., Ou S. I., Going beneath the tip of the iceberg. Identifying and understanding EML4-ALK variants and TP53 mutations to optimize treatment of ALK fusion positive (ALK+) NSCLC. Lung Cancer 158, 126–136 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

