Inferring Pathways of Oxidative Folding from Prefolding Free Energy Landscapes of Disulfide-Rich Toxins
- PMID: 36791259
- PMCID: PMC9987446
- DOI: 10.1021/acs.jpcb.2c07124
Inferring Pathways of Oxidative Folding from Prefolding Free Energy Landscapes of Disulfide-Rich Toxins
Abstract
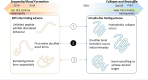
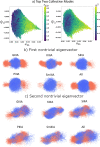
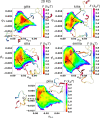
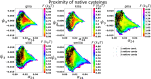
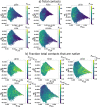
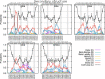
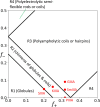
Short, cysteine-rich peptides can exist in stable or metastable structural ensembles due to the number of possible patterns of formation of their disulfide bonds. One interesting subset of this peptide group is the conotoxins, which are produced by aquatic snails in the family Conidae. The μ conotoxins, which are antagonists and blockers of the voltage-gated sodium channel, exist in a folding spectrum: on one end of the spectrum are more hirudin-like folders, which form disulfide bonds and then reshuffle them, leading to an ensemble of kinetically trapped isomers, and on the other end are more BPTI-like folders, which form the native disulfide bonds one by one in a particular order, leading to a preponderance of conformations existing in a single stable state. In this Article, we employ the composite diffusion map approach to study the unified free energy surface of prefolding μ-conotoxin equilibrium. We identify the two most important nonlinear collective modes of the unified folding landscape and demonstrate that in the absence of their disulfides, the conotoxins can be thought of as largely disordered polymers. A small increase in the number of hydrophobic residues in the protein shifts the free energy landscape toward hydrophobically collapsed coil conformations responsible for cysteine proximity in hirudin-like folders, compared to semiextended coil conformations with more distal cysteines in BPTI-like folders. Overall, this work sheds important light on the folding processes and free energy landscapes of cysteine-rich peptides and demonstrates the extent to which sequence and length contribute to these landscapes.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Luo S.; Zhangsun D.; Harvey P. J.; Kaas Q.; Wu Y.; Zhu X.; Hu Y.; Li X.; Tsetlin V. I.; Christensen S.; et al. Cloning, synthesis, and characterization of αO-conotoxin GeXIVA, a potent α9α10 nicotinic acetylcholine receptor antagonist. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E4026–E4035. 10.1073/pnas.1503617112. - DOI - PMC - PubMed
-
- Dang B. Chemical synthesis and structure determination of venom toxins. Chin. Chem. Lett. 2019, 30, 1369–1373. 10.1016/j.cclet.2019.03.021. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

