Mechanistic Insight into the Suppression of Polyglutamine Aggregation by SRCP1
- PMID: 36791332
- PMCID: PMC10023506
- DOI: 10.1021/acschembio.2c00893
Mechanistic Insight into the Suppression of Polyglutamine Aggregation by SRCP1
Abstract
Protein aggregation is a hallmark of the polyglutamine diseases. One potential treatment for these diseases is suppression of polyglutamine aggregation. Previous work identified the cellular slime mold Dictyostelium discoideum as being naturally resistant to polyglutamine aggregation. Further work identified serine-rich chaperone protein 1 (SRCP1) as a protein that is both necessary in Dictyostelium and sufficient in human cells to suppress polyglutamine aggregation. Therefore, understanding how SRCP1 suppresses aggregation may be useful for developing therapeutics for the polyglutamine diseases. Here we utilized a de novo protein modeling approach to generate predictions of SRCP1's structure. Using our best-fit model, we generated mutants that were predicted to alter the stability of SRCP1 and tested these mutants' stability in cells. Using these data, we identified top models of SRCP1's structure that are consistent with the C-terminal region of SRCP1 forming a β-hairpin with a highly dynamic N-terminal region. We next generated a series of peptides that mimic the predicted β-hairpin and validated that they inhibit aggregation of a polyglutamine-expanded mutant huntingtin exon 1 fragment in vitro. To further assess mechanistic details of how SRCP1 inhibits polyglutamine aggregation, we utilized biochemical assays to determine that SRCP1 inhibits secondary nucleation in a manner dependent upon the regions flanking the polyglutamine tract. Finally, to determine if SRCP1 more could generally suppress protein aggregation, we confirmed that it was sufficient to inhibit aggregation of polyglutamine-expanded ataxin-3. Together these studies provide details into the structural and mechanistic basis of the inhibition of protein aggregation by SRCP1.
Figures
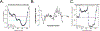

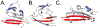





Similar articles
-
SRCP1 Conveys Resistance to Polyglutamine Aggregation.Mol Cell. 2018 Jul 19;71(2):216-228.e7. doi: 10.1016/j.molcel.2018.07.008. Mol Cell. 2018. PMID: 30029002 Free PMC article.
-
The Social Amoeba Dictyostelium discoideum Is Highly Resistant to Polyglutamine Aggregation.J Biol Chem. 2015 Oct 16;290(42):25571-8. doi: 10.1074/jbc.M115.676247. Epub 2015 Sep 1. J Biol Chem. 2015. PMID: 26330554 Free PMC article.
-
Chaperone suppression of cellular toxicity of huntingtin is independent of polyglutamine aggregation.J Biol Chem. 2001 Dec 21;276(51):48417-24. doi: 10.1074/jbc.M104140200. Epub 2001 Oct 17. J Biol Chem. 2001. PMID: 11606565
-
Polyglutamine expansion diseases: More than simple repeats.J Struct Biol. 2018 Feb;201(2):139-154. doi: 10.1016/j.jsb.2017.09.006. Epub 2017 Sep 18. J Struct Biol. 2018. PMID: 28928079 Review.
-
Analyzing the aggregation of polyglutamine-expansion proteins and its modulation by molecular chaperones.Methods. 2011 Mar;53(3):267-74. doi: 10.1016/j.ymeth.2010.12.035. Epub 2010 Dec 30. Methods. 2011. PMID: 21195182 Review.
Cited by
-
Allosteric Modulation of Pathological Ataxin-3 Aggregation: A Path to Spinocerebellar Ataxia Type-3 Therapies.bioRxiv [Preprint]. 2025 Jan 24:2025.01.22.633970. doi: 10.1101/2025.01.22.633970. bioRxiv. 2025. PMID: 39896516 Free PMC article. Preprint.
-
Cell biology of Dictyostelium.BMC Mol Cell Biol. 2025 Aug 19;26(1):25. doi: 10.1186/s12860-025-00550-y. BMC Mol Cell Biol. 2025. PMID: 40830425 Free PMC article.
References
-
- DiFiglia M, Sapp E, Chase K, Davies S, Bates G, Vonsattel J, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–3. - PubMed
-
- Li S, Li X. Aggregation of N-terminal huntingtin is dependent on the length of its glutamine repeats. Hum Mol Genet. 1998;7:777–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

