Nirmatrelvir exerts distinct antiviral potency against different human coronaviruses
- PMID: 36791846
- PMCID: PMC9925195
- DOI: 10.1016/j.antiviral.2023.105555
Nirmatrelvir exerts distinct antiviral potency against different human coronaviruses
Abstract
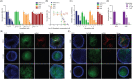
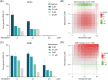
Nirmatrelvir is the main component of Paxlovid, an oral antiviral drug approved for the treatment of COVID-19 caused by SARS-COV-2 infection. Nirmatrelvir targets the main protease (Mpro), which is substantially conserved among different coronaviruses. Here, our molecular docking analysis indicates comparable affinity of nirmatrelvir binding to the Mpro enzymes of SARS-CoV-2 and three seasonal coronaviruses (OC43, 229E and NL63). However, in cell culture models, we found that nirmatrelvir potently inhibited SARS-CoV-2, OC43 and 229E, but not NL63. The insensitivity of NL63 to nirmatrelvir treatment was demonstrated at both viral replication and infectious titer levels. The antiviral activity of nirmatrelvir against OC43 and 229E was further confirmed in human airway organoids. The combination of nirmatrelvir and molnupiravir exerted differential patterns of antiviral response against OC43 and 229E. These results revealed disparities in the ability of nirmatrelvir to inhibit different coronaviruses, and caution against repurposing of nirmatrelvir as a pan-coronavirus treatment.
Keywords: Antiviral activity; Disparity; Nirmatrelvir; Pan-coronavirus.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
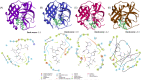
Similar articles
-
Immunosuppressants exert differential effects on pan-coronavirus infection and distinct combinatory antiviral activity with molnupiravir and nirmatrelvir.United European Gastroenterol J. 2023 Jun;11(5):431-447. doi: 10.1002/ueg2.12417. Epub 2023 May 25. United European Gastroenterol J. 2023. PMID: 37226653 Free PMC article.
-
Immunodetection assays for the quantification of seasonal common cold coronaviruses OC43, NL63, or 229E infection confirm nirmatrelvir as broad coronavirus inhibitor.Antiviral Res. 2022 Jul;203:105343. doi: 10.1016/j.antiviral.2022.105343. Epub 2022 May 19. Antiviral Res. 2022. PMID: 35598779 Free PMC article.
-
Recapitulating infection, thermal sensitivity and antiviral treatment of seasonal coronaviruses in human airway organoids.EBioMedicine. 2022 Jul;81:104132. doi: 10.1016/j.ebiom.2022.104132. Epub 2022 Jun 29. EBioMedicine. 2022. PMID: 35779493 Free PMC article.
-
Molnupiravir and Nirmatrelvir-Ritonavir: Oral Coronavirus Disease 2019 Antiviral Drugs.Clin Infect Dis. 2023 Jan 6;76(1):165-171. doi: 10.1093/cid/ciac180. Clin Infect Dis. 2023. PMID: 35245942 Free PMC article. Review.
-
Viral target and metabolism-based rationale for combined use of recently authorized small molecule COVID-19 medicines: Molnupiravir, nirmatrelvir, and remdesivir.Fundam Clin Pharmacol. 2023 Aug;37(4):726-738. doi: 10.1111/fcp.12889. Epub 2023 Mar 25. Fundam Clin Pharmacol. 2023. PMID: 36931725 Free PMC article. Review.
Cited by
-
Characterization of a mouse-adapted strain of bat severe acute respiratory syndrome-related coronavirus.J Virol. 2023 Sep 28;97(9):e0079023. doi: 10.1128/jvi.00790-23. Epub 2023 Aug 21. J Virol. 2023. PMID: 37607058 Free PMC article.
-
Drug-Drug Interactions Leading to Tacrolimus Toxicity in a Renal Transplant Patient With COVID-19: The Role of Paxlovid and the Mitigating Use of Phenytoin.Cureus. 2025 Mar 20;17(3):e80902. doi: 10.7759/cureus.80902. eCollection 2025 Mar. Cureus. 2025. PMID: 40255720 Free PMC article.
-
Potential Broad-Spectrum Antiviral Agents: A Key Arsenal Against Newly Emerging and Reemerging Respiratory RNA Viruses.Int J Mol Sci. 2025 Feb 10;26(4):1481. doi: 10.3390/ijms26041481. Int J Mol Sci. 2025. PMID: 40003946 Free PMC article. Review.
-
A low-background, fluorescent assay to evaluate inhibitors of diverse viral proteases.J Virol. 2023 Aug 31;97(8):e0059723. doi: 10.1128/jvi.00597-23. Epub 2023 Aug 14. J Virol. 2023. PMID: 37578235 Free PMC article.
-
Seasonal human coronaviruses OC43, 229E, and NL63 induce cell surface modulation of entry receptors and display host cell-specific viral replication kinetics.Microbiol Spectr. 2024 Jul 2;12(7):e0422023. doi: 10.1128/spectrum.04220-23. Epub 2024 Jun 12. Microbiol Spectr. 2024. PMID: 38864599 Free PMC article.
References
-
- Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. - PMC - PubMed
-
- Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. - PubMed
-
- Han Y., Yang L., Lacko L.A., Chen S. Human organoid models to study SARS-CoV-2 infection. Nat. Methods. 2022;19:418–428. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

