Distal Fecal Wash Host Transcriptomics Identifies Inflammation Throughout the Colon and Terminal Ileum
- PMID: 36791991
- PMCID: PMC10199420
- DOI: 10.1016/j.jcmgh.2023.02.002
Distal Fecal Wash Host Transcriptomics Identifies Inflammation Throughout the Colon and Terminal Ileum
Abstract
Background & aims: Noninvasive modalities for assessing active endoscopic and histologic inflammation in Crohn's disease and ulcerative colitis patients are critically needed. Fecal wash host shed-cell transcriptomics has been shown to be a robust classifier of endoscopic and histologic inflammation in inflammatory bowel disease patients with distal colitis. Whether such fecal washes can inform on inflammatory processes occurring in more proximal intestinal segments is currently unknown.
Methods: Fifty-nine inflammatory bowel disease patients and 50 controls were prospectively enrolled. Biopsy specimens and fecal washes from the distal colon, proximal colon, and terminal ileum were compared. Host transcriptomics were performed on the biopsy specimens and fecal washes obtained during colonoscopy at predefined locations throughout the colon and terminal ileum and results were associated with concurrent clinical, endoscopic, and histologic parameters.
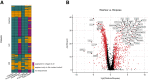
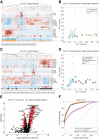
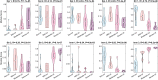
Results: We found that host transcriptomics of distal fecal washes robustly classify histologic inflammation in ileal and proximal colonic Crohn's disease, even without distal colonic involvement (area under the receiver operating characteristic curve, 0.94 ± 0.09). We further found that fecal washes consist of modules of co-expressed genes of immune, stromal, and epithelial origin that are indicative of endoscopic disease severity. Fecal wash host transcriptomics also captures expression of gene modules previously associated with a lack of response to biological therapies.
Conclusions: Our study establishes the accuracy of distal colonic fecal washes for identifying and scoring inflammatory processes throughout the entire ileal-colonic axis.
Keywords: Histology; IBD; Therapy Outcome; Transcriptomics.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
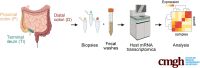





Comment in
-
Distal Fecal Wash: Intestinal Liquid Biopsy?Cell Mol Gastroenterol Hepatol. 2023;16(1):161-162. doi: 10.1016/j.jcmgh.2023.04.001. Epub 2023 Apr 29. Cell Mol Gastroenterol Hepatol. 2023. PMID: 37130565 Free PMC article. No abstract available.
References
-
- Ungar B., Yavzori M., Fudim E., et al. Host transcriptome signatures in human faecal-washes predict histological remission in patients with IBD. Gut. 2022;71:1988–1997. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

