Personalized recurrence risk assessment following the birth of a child with a pathogenic de novo mutation
- PMID: 36792598
- PMCID: PMC9932158
- DOI: 10.1038/s41467-023-36606-w
Personalized recurrence risk assessment following the birth of a child with a pathogenic de novo mutation
Abstract
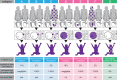
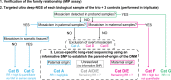
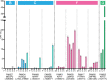
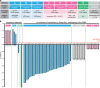
Following the diagnosis of a paediatric disorder caused by an apparently de novo mutation, a recurrence risk of 1-2% is frequently quoted due to the possibility of parental germline mosaicism; but for any specific couple, this figure is usually incorrect. We present a systematic approach to providing individualized recurrence risk. By combining locus-specific sequencing of multiple tissues to detect occult mosaicism with long-read sequencing to determine the parent-of-origin of the mutation, we show that we can stratify the majority of couples into one of seven discrete categories associated with substantially different risks to future offspring. Among 58 families with a single affected offspring (representing 59 de novo mutations in 49 genes), the recurrence risk for 35 (59%) was decreased below 0.1%, but increased owing to parental mixed mosaicism for 5 (9%)-that could be quantified in semen for paternal cases (recurrence risks of 5.6-12.1%). Implementation of this strategy offers the prospect of driving a major transformation in the practice of genetic counselling.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

