Molecular mechanisms of coronary artery disease risk at the PDGFD locus
- PMID: 36792607
- PMCID: PMC9932166
- DOI: 10.1038/s41467-023-36518-9
Molecular mechanisms of coronary artery disease risk at the PDGFD locus
Abstract
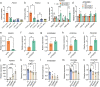
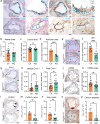
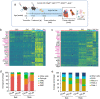
Genome wide association studies for coronary artery disease (CAD) have identified a risk locus at 11q22.3. Here, we verify with mechanistic studies that rs2019090 and PDGFD represent the functional variant and gene at this locus. Further, FOXC1/C2 transcription factor binding at rs2019090 is shown to promote PDGFD transcription through the CAD promoting allele. With single cell transcriptomic and histology studies with Pdgfd knockdown in an SMC lineage tracing male atherosclerosis mouse model we find that Pdgfd promotes expansion, migration, and transition of SMC lineage cells to the chondromyocyte phenotype. Pdgfd also increases adventitial fibroblast and pericyte expression of chemokines and leukocyte adhesion molecules, which is linked to plaque macrophage recruitment. Despite these changes there is no effect of Pdgfd deletion on overall plaque burden. These findings suggest that PDGFD mediates CAD risk by promoting deleterious phenotypic changes in SMC, along with an inflammatory response that is primarily focused in the adventitia.
© 2023. The Author(s).
Conflict of interest statement
C.S.W. is a consultant for Tensixteen Bio and Renovacor, T.Q. is a consultant for Saliogen, and a member of the Cardiometabolic Scientific Advisory Board of Amgen. S.J. and S.H. are paid employees of Amgen, and this work was supported in part by Amgen, Inc. All other authors declare no competing interests
Figures



Update of
-
Molecular mechanisms of coronary artery disease risk at the PDGFD locus.bioRxiv [Preprint]. 2023 Jan 27:2023.01.26.525789. doi: 10.1101/2023.01.26.525789. bioRxiv. 2023. Update in: Nat Commun. 2023 Feb 15;14(1):847. doi: 10.1038/s41467-023-36518-9. PMID: 36747745 Free PMC article. Updated. Preprint.
References
-
- Erdmann J, Kessler T, Munoz Venegas L, Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc. Res. 2018;114:1241–1257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

