Generalisability of fetal ultrasound deep learning models to low-resource imaging settings in five African countries
- PMID: 36792642
- PMCID: PMC9932015
- DOI: 10.1038/s41598-023-29490-3
Generalisability of fetal ultrasound deep learning models to low-resource imaging settings in five African countries
Erratum in
-
Author Correction: Generalisability of fetal ultrasound deep learning models to low-resource imaging settings in five African countries.Sci Rep. 2023 Feb 28;13(1):3410. doi: 10.1038/s41598-023-30540-z. Sci Rep. 2023. PMID: 36854807 Free PMC article. No abstract available.
Abstract
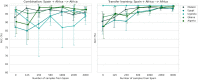
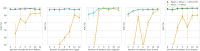
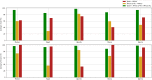
Most artificial intelligence (AI) research and innovations have concentrated in high-income countries, where imaging data, IT infrastructures and clinical expertise are plentiful. However, slower progress has been made in limited-resource environments where medical imaging is needed. For example, in Sub-Saharan Africa, the rate of perinatal mortality is very high due to limited access to antenatal screening. In these countries, AI models could be implemented to help clinicians acquire fetal ultrasound planes for the diagnosis of fetal abnormalities. So far, deep learning models have been proposed to identify standard fetal planes, but there is no evidence of their ability to generalise in centres with low resources, i.e. with limited access to high-end ultrasound equipment and ultrasound data. This work investigates for the first time different strategies to reduce the domain-shift effect arising from a fetal plane classification model trained on one clinical centre with high-resource settings and transferred to a new centre with low-resource settings. To that end, a classifier trained with 1792 patients from Spain is first evaluated on a new centre in Denmark in optimal conditions with 1008 patients and is later optimised to reach the same performance in five African centres (Egypt, Algeria, Uganda, Ghana and Malawi) with 25 patients each. The results show that a transfer learning approach for domain adaptation can be a solution to integrate small-size African samples with existing large-scale databases in developed countries. In particular, the model can be re-aligned and optimised to boost the performance on African populations by increasing the recall to [Formula: see text] and at the same time maintaining a high precision across centres. This framework shows promise for building new AI models generalisable across clinical centres with limited data acquired in challenging and heterogeneous conditions and calls for further research to develop new solutions for the usability of AI in countries with fewer resources and, consequently, in higher need of clinical support.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Houssein EH, Emam MM, Ali AA, Suganthan PN. Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review. Expert Syst. Appl. 2021;167:114161. doi: 10.1016/j.eswa.2020.114161. - DOI
-
- Zhou, S. K., Greenspan, H., Davatzikos, C., Duncan, J. S., Van Ginneken, B., Madabhushi, A., Prince, J. L., Rueckert, D., & Summers, R. M. A review of deep learning in medical imaging: Imaging traits, technology trends, case studies with progress highlights, and future promises. In Proceedings of the IEEE (2021). - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

