Functional connectivity signatures of major depressive disorder: machine learning analysis of two multicenter neuroimaging studies
- PMID: 36792654
- PMCID: PMC10615764
- DOI: 10.1038/s41380-023-01977-5
Functional connectivity signatures of major depressive disorder: machine learning analysis of two multicenter neuroimaging studies
Abstract
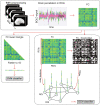
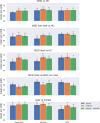
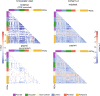
The promise of machine learning has fueled the hope for developing diagnostic tools for psychiatry. Initial studies showed high accuracy for the identification of major depressive disorder (MDD) with resting-state connectivity, but progress has been hampered by the absence of large datasets. Here we used regular machine learning and advanced deep learning algorithms to differentiate patients with MDD from healthy controls and identify neurophysiological signatures of depression in two of the largest resting-state datasets for MDD. We obtained resting-state functional magnetic resonance imaging data from the REST-meta-MDD (N = 2338) and PsyMRI (N = 1039) consortia. Classification of functional connectivity matrices was done using support vector machines (SVM) and graph convolutional neural networks (GCN), and performance was evaluated using 5-fold cross-validation. Features were visualized using GCN-Explainer, an ablation study and univariate t-testing. The results showed a mean classification accuracy of 61% for MDD versus controls. Mean accuracy for classifying (non-)medicated subgroups was 62%. Sex classification accuracy was substantially better across datasets (73-81%). Visualization of the results showed that classifications were driven by stronger thalamic connections in both datasets, while nearly all other connections were weaker with small univariate effect sizes. These results suggest that whole brain resting-state connectivity is a reliable though poor biomarker for MDD, presumably due to disease heterogeneity as further supported by the higher accuracy for sex classification using the same methods. Deep learning revealed thalamic hyperconnectivity as a prominent neurophysiological signature of depression in both multicenter studies, which may guide the development of biomarkers in future studies.
© 2023. The Author(s).
Conflict of interest statement
GvW received research funding from Philips. The other authors declare no competing interests.
Figures

References
-
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–858. doi: 10.1016/S0140-6736(18)32279-7. - DOI - PMC - PubMed
-
- Coleman JRI, Gaspar HA, Bryois J, Bipolar Disorder Working Group of the Psychiatric Genomics Consortium, Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Breen G. The genetics of the mood disorder spectrum: genome-wide association analyses of more than 185,000 cases and 439,000 controls. Biol Psychiatry. 2020;88:169–84. doi: 10.1016/j.biopsych.2019.10.015. - DOI - PMC - PubMed

