Selectivity for food in human ventral visual cortex
- PMID: 36792693
- PMCID: PMC9932019
- DOI: 10.1038/s42003-023-04546-2
Selectivity for food in human ventral visual cortex
Abstract
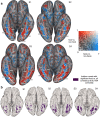
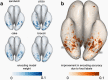
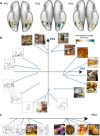
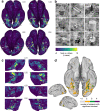
Visual cortex contains regions of selectivity for domains of ecological importance. Food is an evolutionarily critical category whose visual heterogeneity may make the identification of selectivity more challenging. We investigate neural responsiveness to food using natural images combined with large-scale human fMRI. Leveraging the improved sensitivity of modern designs and statistical analyses, we identify two food-selective regions in the ventral visual cortex. Our results are robust across 8 subjects from the Natural Scenes Dataset (NSD), multiple independent image sets and multiple analysis methods. We then test our findings of food selectivity in an fMRI "localizer" using grayscale food images. These independent results confirm the existence of food selectivity in ventral visual cortex and help illuminate why earlier studies may have failed to do so. Our identification of food-selective regions stands alongside prior findings of functional selectivity and adds to our understanding of the organization of knowledge within the human visual system.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

