Identification of the global diurnal rhythmic transcripts, transcription factors and time-of-day specific cis elements in Chenopodium quinoa
- PMID: 36793005
- PMCID: PMC9933291
- DOI: 10.1186/s12870-023-04107-z
Identification of the global diurnal rhythmic transcripts, transcription factors and time-of-day specific cis elements in Chenopodium quinoa
Abstract
Background: Photoperiod is an important environmental cue interacting with circadian clock pathway to optimize the local adaption and yield of crops. Quinoa (Chenopodium quinoa) in family Amaranthaceae has been known as superfood due to the nutritious elements. As quinoa was originated from the low-latitude Andes, most of the quinoa accessions are short-day type. Short-day type quinoa usually displays altered growth and yield status when introduced into higher latitude regions. Thus, deciphering the photoperiodic regulation on circadian clock pathway will help breed adaptable and high yielding quinoa cultivars.
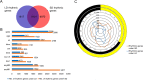
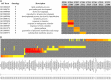
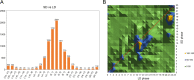
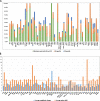
Results: In this study, we conducted RNA-seq analysis of the diurnally collected leaves of quinoa plants treated by short-day (SD) and long-day conditions (LD), respectively. We identified 19,818 (44% of global genes) rhythmic genes in quinoa using HAYSTACK analysis. We identified the putative circadian clock architecture and investigated the photoperiodic regulatory effects on the expression phase and amplitude of global rhythmic genes, core clock components and transcription factors. The global rhythmic transcripts were involved in time-of-day specific biological processes. A higher percentage of rhythmic genes had advanced phases and strengthened amplitudes when switched from LD to SD. The transcription factors of CO-like, DBB, EIL, ERF, NAC, TALE and WRKY families were sensitive to the day length changes. We speculated that those transcription factors may function as key mediators for the circadian clock output in quinoa. Besides, we identified 15 novel time-of-day specific motifs that may be key cis elements for rhythm-keeping in quinoa.
Conclusions: Collectively, this study lays a foundation for understanding the circadian clock pathway and provides useful molecular resources for adaptable elites breeding in quinoa.
Keywords: Chenopodium quinoa; Cis elements; Diurnal rhythmic genes; Phase shift; Photoperiod; Transcription factors.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Investigation into the underlying regulatory mechanisms shaping inflorescence architecture in Chenopodium quinoa.BMC Genomics. 2019 Aug 17;20(1):658. doi: 10.1186/s12864-019-6027-0. BMC Genomics. 2019. PMID: 31419932 Free PMC article.
-
Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules.PLoS One. 2011;6(6):e16907. doi: 10.1371/journal.pone.0016907. Epub 2011 Jun 9. PLoS One. 2011. PMID: 21694767 Free PMC article.
-
Leaf and shoot apical meristem transcriptomes of quinoa (Chenopodium quinoa Willd.) in response to photoperiod and plant development.Plant Cell Environ. 2024 Jun;47(6):2027-2043. doi: 10.1111/pce.14864. Epub 2024 Feb 23. Plant Cell Environ. 2024. PMID: 38391415
-
From a repressilator-based circadian clock mechanism to an external coincidence model responsible for photoperiod and temperature control of plant architecture in Arabodopsis thaliana.Biosci Biotechnol Biochem. 2013;77(1):10-6. doi: 10.1271/bbb.120765. Epub 2013 Jan 7. Biosci Biotechnol Biochem. 2013. PMID: 23291766 Review.
-
Regulatory principles of photoperiod-driven clock function in plants.Trends Plant Sci. 2025 Jun;30(6):594-602. doi: 10.1016/j.tplants.2025.01.008. Epub 2025 Feb 20. Trends Plant Sci. 2025. PMID: 39984377 Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

