Clinical utility of genomic profiling of AML using paraffin-embedded bone marrow clots: HM-SCREEN-Japan 01
- PMID: 36793248
- PMCID: PMC10154825
- DOI: 10.1111/cas.15746
Clinical utility of genomic profiling of AML using paraffin-embedded bone marrow clots: HM-SCREEN-Japan 01
Abstract
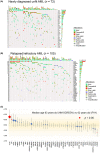
Next-generation sequencing of AML has identified specific genetic mutations in AML patients. Hematologic Malignancies (HM)-SCREEN-Japan 01 is a multicenter study to detect actionable mutations using paraffin-embedded bone marrow (BM) clot specimens rather than BM fluid in AML patients for whom standard treatment has not been established. The purpose of this study is to evaluate the presence of potentially therapeutic target gene mutations in patients with newly diagnosed unfit AML and relapsed/refractory AML (R/R-AML) using BM clot specimens. In this study, 188 patients were enrolled and targeted sequencing was undertaken on DNA from 437 genes and RNA from 265 genes. High-quality DNA and RNA were obtained using BM clot specimens, with genetic alterations successfully detected in 177 patients (97.3%), and fusion transcripts in 41 patients (23.2%). The median turnaround time was 13 days. In the detection of fusion genes, not only common fusion products such as RUNX1-RUX1T1 and KMT2A rearrangements, but also NUP98 rearrangements and rare fusion genes were observed. Among 177 patients (72 with unfit AML, 105 with R/R-AML), mutations in KIT and WT1 were independent factors for overall survival (hazard ratio = 12.6 and 8.88, respectively), and patients with high variant allele frequency (≥40%) of TP53 mutations had a poor prognosis. As for the detection of actionable mutations, 38% (n = 69) of patients had useful genetic mutation (FLT3-ITD/TKD, IDH1/2, and DNMT3AR822 ) for treatment selection. Comprehensive genomic profiling using paraffin-embedded BM clot specimens successfully identified leukemic-associated genes that can be used as therapeutic targets.
Keywords: AML; FoundationOne Heme; actionable mutation; ineligible for standard chemotherapy; paraffin-embedded bone marrow clot.
© 2023 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
T.Y.: Otsuka, Pfizer, Abbie, Astellas, Daiichi Sankyo, Solasia Pharma (research funding); Ono Pharmaceutical, Pfizer, Chugai (honoraria). Y.M.: Bristol‐Myers Squibb, Novartis Pharma KK, Pfizer Japan, Inc., Takeda (honoraria). H.S.: Astellas, Teijin, Shionogi, Taiho, Eisai, Celgene, Ono, Takeda, Merck Sharp & Dohme, Sumitomo Dainippon, Nippon Shinyaku, Novartis, Janssen, Chugai, AbbVie (research funding); Eisai, Ono, Takeda, Sumitomo Dainippon, Nippon Shinyaku, Daiichi Sankyo, Novartis, Janssen, Chugai, Kyowa Kirin, Otsuka, Bristol‐Myers Squibb, Pfizer, Fujimoto, AbbVie, AstraZeneca, Sanofi, Mundi Pharma (honoraria); Eisai, Celgene, Chugai, AbbVie, AstraZeneca (membership on an entity's Board of Directors or advisory committees). K.Y.: AbbVie, Astra‐Zeneca, Bayer, Celgene, Chugai, Eisai, IQIVA/Incyte, Gilead Sciences, MSD, Mundipharma, Nippon Shinyaku, Novartis, Ono, Otsuka, Solasia Pharma, SymBio, Takeda, Yakult, Zenyaku (research funding); AbbVie, Bristol‐Myers Squibb, Celgene, Chugai, Eisai, IQIVA/HUYA, Janssen, Kyowa Kirin, Meiji Seika Pharma, Mochida, MSD, Mundipharma, Nippon Shinyaku, Novartis, Ono, Otsuka, Pfizer, Sanofi, Sumitomo Dainippon, Takeda (honoraria); AbbVie, Astra‐Zeneca, Celgene, Chugai, Eisai, Daiichi Sankyo, HUYA, Meiji Seika Pharma, MSD, Mundipharma, Ono, Otsuka, Stemline Therapeutics, Takeda (consultancy). J.K.: Bristol‐Myers Squibb, Chugai Pharmaceutical, Dainippon Sumitomo Pharma, Daiichi Sankyo, Sanofi, Kyowa Kirin, Otsuka Pharmaceutical, Astellas Pharma, Takeda, Celgene, MSD, Ono Pharmaceutical, Eisai, Sysmex, Pfizer, Nippon Shinyaku, Shionogi, Asahi Kasei, Taiho Pharmaceutical, Fujimoto Pharmaceutical (research funding); Bristol‐Myers Squibb, Chugai Pharmaceutical, Dainippon Sumitomo Pharma, Daiichi Sankyo, Sanofi, Kyowa Kirin, Otsuka Pharmaceutical, Astellas Pharma, Takeda, Celgene, Abbvie, Ono Pharmaceutical, Eisai, Pfizer, Nippon Shinyaku, Fujimoto Pharmaceutical (honoraria); Janssen Pharmaceutical KK, Bristol‐Myers Squibb, Sanofi, Celgene, Abbvie (consultancy). K.U.: Astellas, Abbvie, Gilead, Symbio, Daiichi Sankyo, Sumitomo Dainippon, Otsuka, Novartis, Bristol‐Myers Squibb, Ono, Janssen, Celgene, Takeda, Nippon Boehringer Ingelheim, Mundipharma, Astellas‐Amgen‐Biopharma, Nippon Shinyaku, Kyowa Kirin, Pfizer (research funding); Astellas, Symbio, Daiichi Sankyo, Otsuka, Novartis, Bristol‐Myers Squibb, Ono, Celgene, Nippon Shinyaku, Kyowa Kirin, Alexion, Eisai, MSD, Takeda, PharmaEssentia, Yakult (speakers bureau). A.G.: Eisai Co., Ltd., Ono Pharmaceutical Co., Ltd., Taiho Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., Nippon Shinyaku Co., Ltd., Chugai Pharmaceutical Co., Ltd., MSD KK, Otsuka Pharmaceutical Co., Ltd., Sumitomo Dainippon Pharma Co., Ltd., Bayer Yakuhin, Ltd., Daiichi‐Sankyo Co., Ltd., and Nihon Pharmaceutical Co., Ltd. (research funding); Novartis Pharma KK, Alexion Pharmaceuticals, Inc., Eisai Co., Ltd., Ono Pharmaceutical Co., Ltd., Taiho Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., Nippon Shinyaku Co., Ltd., Chugai Pharmaceutical Co., Ltd., Otsuka Pharmaceutical Co., Ltd., Sumitomo Dainippon Pharma Co., Ltd., Daiichi‐Sankyo Co., Ltd., Nihon Pharmaceutical Co., Ltd., Kyowa Kirin Co., Ltd., Janssen Pharmaceutical KK, Pfizer Japan Inc., Sanofi KK (honoraria); PharmaEssentia Japan KK, Chugai Pharmaceutical Co. (consultancy).
Figures



References
-
- Perl AE, Martinelli G, Cortes JE, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3‐mutated AML. N Engl J Med. 2019;381(18):1728‐1740. - PubMed
-
- DiNardo CD, Stein EM, de Botton S, et al. Durable remissions with Ivosidenib in IDH1‐mutated relapsed or refractory AML. N Engl J Med. 2018;378(25):2386‐2398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

