Identification of Differentially Methylated Genes Associated with Clear Cell Renal Cell Carcinoma and Their Prognostic Values
- PMID: 36793506
- PMCID: PMC9925242
- DOI: 10.1155/2023/8405945
Identification of Differentially Methylated Genes Associated with Clear Cell Renal Cell Carcinoma and Their Prognostic Values
Retraction in
-
Retracted: Identification of Differentially Methylated Genes Associated with Clear Cell Renal Cell Carcinoma and Their Prognostic Values.J Environ Public Health. 2023 Dec 13;2023:9756067. doi: 10.1155/2023/9756067. eCollection 2023. J Environ Public Health. 2023. PMID: 38125879 Free PMC article.
Abstract
Objective: Renal cell carcinoma (RCC) is a heterogeneous disease comprising histologically defined subtypes among which clear cell RCC (ccRCC) accounts for 70% of all RCC cases. DNA methylation constitutes a main part of the molecular mechanism of cancer evolution and prognosis. In this study, we aim to identify differentially methylated genes related to ccRCC and their prognostic values.
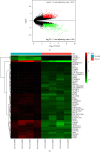
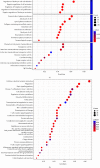
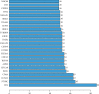
Methods: The GSE168845 dataset was downloaded from the Gene Expression Omnibus (GEO) database to identify differentially expressed genes (DEGs) between ccRCC tissues and paired tumor-free kidney tissues. DEGs were submitted to public databases for functional and pathway enrichment analysis, protein-protein interaction (PPI) analysis, promoter methylation analysis, and survival correlation analysis.
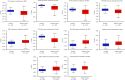
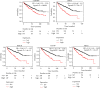
Results: In the setting of |log2FC| ≥ 2 and adjusted p value <0.05 during differential expression analysis of the GSE168845 dataset, 1659 DEGs between ccRCC tissues and paired tumor-free kidney tissues were sorted out. The most enriched pathways were "T cell activation" and "cytokine-cytokine receptor interaction." After PPI analysis, 22 hub genes related to ccRCC stood out, among which CD4, PTPRC, ITGB2, TYROBP, BIRC5, and ITGAM exhibited higher methylation levels, and BUB1B, CENPF, KIF2C, and MELK exhibited lower methylation levels in ccRCC tissues compared with paired tumor-free kidney tissues. Among these differentially methylated genes, TYROBP, BIRC5, BUB1B, CENPF, and MELK were significantly correlated with the survival of ccRCC patients (p < 0.001).
Conclusion: Our study indicates the DNA methylation of TYROBP, BIRC5, BUB1B, CENPF, and MELK may be promising results for the prognosis of ccRCC.
Copyright © 2023 Bin Wan et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
References
-
- Sung H., Ferlay J., Siegel R. L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians . 2021;71(3):209–249. - PubMed
-
- Bukavina L., Bensalah K., Bray F., et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eurpean Urology . 2022;82(5) - PubMed
-
- Powles T., Rini B. Novel agents and drug development needs in advanced clear cell renal cancer. Journal of Clinical Oncology . 2018;79:p. 2655. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

