Hyperactive neuronal autophagy depletes BDNF and impairs adult hippocampal neurogenesis in a corticosterone-induced mouse model of depression
- PMID: 36793868
- PMCID: PMC9925310
- DOI: 10.7150/thno.81067
Hyperactive neuronal autophagy depletes BDNF and impairs adult hippocampal neurogenesis in a corticosterone-induced mouse model of depression
Abstract
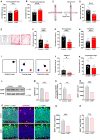
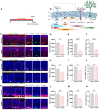
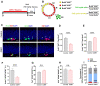
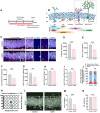
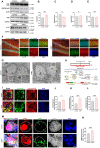
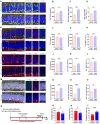
Background: Depression is a mental disorder that poses a serious threat to human health. Adult hippocampal neurogenesis (AHN) is closely associated with the efficacy of antidepressants. Chronic treatment with corticosterone (CORT), a well-validated pharmacological stressor, induces depressive-like behaviors and suppresses AHN in experimental animals. However, the possible mechanisms of chronic CORT action remain elusive. Methods: A chronic CORT treatment (0.1 mg/mL, drinking water for 4 weeks) was applied to prepare a mouse model of depression. Immunofluorescence was performed to analyze the hippocampal neurogenesis lineage, and immunoblotting, immunofluorescence, electron microscopy, and adeno-associated virus (AAV) expressing a pH-sensitive tandemly tagged light chain 3 (LC3) protein were used to analyze neuronal autophagy. AAV-hSyn-miR30-shRNA was used to knock down autophagy-related gene 5 (Atg5) expression in the neurons. Results: Chronic CORT induces depressive-like behaviors and decreases the expression of neuronal brain-derived neurotrophic factor (BDNF) in the dentate gyrus (DG) of the hippocampus in mice. Moreover, it markedly diminishes the proliferation of neural stem cells (NSCs), neural progenitor cells, and neuroblasts and impairs the survival and migration of newborn immature and mature neurons in the DG, which may be attributed to changes in the cell cycle kinetics and induction of NSCs apoptosis. Furthermore, chronic CORT induces hyperactive neuronal autophagy in the DG, possibly by increasing the expression of ATG5 and causing excess lysosomal degradation of BDNF in neurons. Notably, inhibiting hyperactive neuronal autophagy in the DG of mice by knocking down Atg5 in neurons using RNA interference reverses the decrease of neuronal BDNF expression, rescues AHN, and exerts antidepressant effects. Conclusion: Our findings reveal a neuronal autophagy-dependent mechanism that links chronic CORT to reduced neuronal BDNF levels, AHN suppression and depressive-like behavior in mice. In addition, our results provide insights for treating depression by targeting neuronal autophagy in the DG of the hippocampus.
Keywords: Adult hippocampal neurogenesis; BDNF; Corticosterone; Depression; Neuronal autophagy..
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

Similar articles
-
Effects of corticosterone on the expression of mature brain-derived neurotrophic factor (mBDNF) and proBDNF in the hippocampal dentate gyrus.Behav Brain Res. 2019 Jun 3;365:150-156. doi: 10.1016/j.bbr.2019.03.010. Epub 2019 Mar 6. Behav Brain Res. 2019. PMID: 30851317
-
Autophagic death of neural stem cells mediates chronic stress-induced decline of adult hippocampal neurogenesis and cognitive deficits.Autophagy. 2020 Mar;16(3):512-530. doi: 10.1080/15548627.2019.1630222. Epub 2019 Jun 24. Autophagy. 2020. PMID: 31234698 Free PMC article.
-
The leptin/BDNF/TrkB signaling pathway improves corticosteroid combined with chronic restraint stress-induced depressive-like behavior in mice.Neurosci Lett. 2025 Jul 13;859-861:138268. doi: 10.1016/j.neulet.2025.138268. Epub 2025 May 17. Neurosci Lett. 2025. PMID: 40389053
-
Roles of microglia in adult hippocampal neurogenesis in depression and their therapeutics.Front Immunol. 2023 Oct 10;14:1193053. doi: 10.3389/fimmu.2023.1193053. eCollection 2023. Front Immunol. 2023. PMID: 37881439 Free PMC article. Review.
-
Methods to study adult hippocampal neurogenesis in humans and across the phylogeny.Hippocampus. 2023 Apr;33(4):271-306. doi: 10.1002/hipo.23474. Epub 2022 Oct 18. Hippocampus. 2023. PMID: 36259116 Free PMC article. Review.
Cited by
-
Hippocampal Neurogenesis in Alzheimer's Disease: Multimodal Therapeutics and the Neurogenic Impairment Index Framework.Int J Mol Sci. 2025 Jun 25;26(13):6105. doi: 10.3390/ijms26136105. Int J Mol Sci. 2025. PMID: 40649882 Free PMC article. Review.
-
Dual-factor model of sleep and diet: a new approach to understanding central fatigue.Front Neurosci. 2024 Sep 17;18:1465568. doi: 10.3389/fnins.2024.1465568. eCollection 2024. Front Neurosci. 2024. PMID: 39355851 Free PMC article.
-
Integrated multi-omics profiling highlights the diet-gut-brain axis in low-calorie diets promoted novelty-seeking behavior.Curr Res Food Sci. 2024 Oct 28;9:100897. doi: 10.1016/j.crfs.2024.100897. eCollection 2024. Curr Res Food Sci. 2024. PMID: 39555017 Free PMC article.
-
The Antidepressant Effect of Magnolol on Depression-Like Behavior of CORT-Treated Mice.J Mol Neurosci. 2024 Jan 6;74(1):3. doi: 10.1007/s12031-023-02185-0. J Mol Neurosci. 2024. PMID: 38183534
-
A vascular endothelial cell, neuron, and microglia tri-culture model to study hypertension-related depression.Front Cell Neurosci. 2025 Mar 31;19:1553309. doi: 10.3389/fncel.2025.1553309. eCollection 2025. Front Cell Neurosci. 2025. PMID: 40230380 Free PMC article.
References
-
- Dubovsky SL, Ghosh BM, Serotte JC, Cranwell V. Psychotic depression: diagnosis, differential diagnosis, and treatment. Psychother Psychosom. 2021;90:160–77. - PubMed
-
- Cruz-Pereira JS, Rea K, Nolan YM, O'Leary OF, Dinan TG, Cryan JF. Depression's unholy trinity: dysregulated stress, immunity, and the microbiome. Annu Rev Psychol. 2020;71:49–78. - PubMed
-
- de Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–75. - PubMed
-
- Dean J, Keshavan M. The neurobiology of depression: an integrated view. Asian J Psychiatr. 2017;27:101–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

