Emergence and pandemic spread of small ruminant lentiviruses
- PMID: 36793939
- PMCID: PMC9924038
- DOI: 10.1093/ve/vead005
Emergence and pandemic spread of small ruminant lentiviruses
Abstract
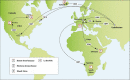
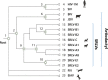
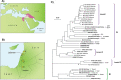
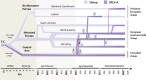
Small ruminant lentiviruses (SRLVs) cause chronic, persistent infections in populations of domestic sheep (Ovis aries) and goats (Capra hircus) worldwide. The vast majority of SRLV infections involve two genotypes (A and B) that spread in association with the emergence of global livestock trade. However, SRLVs have likely been present in Eurasian ruminant populations since at least the early Neolithic period. Here, we use phylogenetic and phylogeographic approaches to reconstruct the origin of pandemic SRLV strains and infer their historical pattern of global spread. We constructed an open computational resource ('Lentivirus-GLUE') via which an up-to-date database of published SRLV sequences, multiple sequence alignments (MSAs), and sequence-associated metadata can be maintained. We used data collated in Lentivirus-GLUE to perform a comprehensive phylogenetic investigation of global SRLV diversity. Phylogenies reconstructed from genome-length alignments reveal that the deep divisions in the SRLV phylogeny are consistent with an ancient split into Eastern (A-like) and Western (B-like) lineages as agricultural systems disseminated out of domestication centres during the Neolithic period. These findings are also consistent with historical and phylogeographic evidence linking the early 20th century emergence of SRLV-A to the international export of Central Asian Karakul sheep. Investigating the global diversity of SRLVs can help reveal how anthropogenic factors have impacted the ecology and evolution of livestock diseases. The open resources generated in our study can expedite these studies and can also serve more broadly to facilitate the use of genomic data in SRLV diagnostics and research.
Keywords: emergence; evolution; iatrogenesis; lentivirus; retrovirus.
© The Author(s) 2023. Published by Oxford University Press.
Figures

Similar articles
-
Genetic characterization of small ruminant lentiviruses circulating in naturally infected sheep and goats in Ontario, Canada.Virus Res. 2013 Jul;175(1):30-44. doi: 10.1016/j.virusres.2013.03.019. Epub 2013 Apr 10. Virus Res. 2013. PMID: 23583225
-
Phylogenetic analysis of small ruminant lentiviruses in Germany and Iran suggests their expansion with domestic sheep.Sci Rep. 2020 Feb 10;10(1):2243. doi: 10.1038/s41598-020-58990-9. Sci Rep. 2020. PMID: 32042070 Free PMC article.
-
Small ruminant lentiviruses (SRLVs) break the species barrier to acquire new host range.Viruses. 2013 Jul 23;5(7):1867-84. doi: 10.3390/v5071867. Viruses. 2013. PMID: 23881276 Free PMC article. Review.
-
Molecular Characterization of Small Ruminant Lentiviruses Isolated from Polish Goats with Arthritis.Viruses. 2022 Mar 31;14(4):735. doi: 10.3390/v14040735. Viruses. 2022. PMID: 35458465 Free PMC article.
-
Molecular Characterization of Small Ruminant Lentiviruses in Sheep and Goats: A Systematic Review.Animals (Basel). 2024 Dec 8;14(23):3545. doi: 10.3390/ani14233545. Animals (Basel). 2024. PMID: 39682510 Free PMC article. Review.
Cited by
-
Serological and Molecular Characterization of Small Ruminant Lentiviruses in Morocco.Animals (Basel). 2024 Feb 7;14(4):550. doi: 10.3390/ani14040550. Animals (Basel). 2024. PMID: 38396519 Free PMC article.
-
Testing the Tenacity of Small Ruminant Lentiviruses In Vitro to Assess the Potential Risk of Indirect Fomites' Transmission.Viruses. 2025 Mar 14;17(3):419. doi: 10.3390/v17030419. Viruses. 2025. PMID: 40143344 Free PMC article.
-
A Combined Approach for the Characterization of Small Ruminant Lentivirus Strains Circulating in the Islands and Mainland of Greece.Animals (Basel). 2024 Apr 6;14(7):1119. doi: 10.3390/ani14071119. Animals (Basel). 2024. PMID: 38612358 Free PMC article.
-
Isolation and Identification of Caprine Arthritis Encephalitis Virus from Animals in the Republic of Mordovia.Animals (Basel). 2023 Jul 13;13(14):2290. doi: 10.3390/ani13142290. Animals (Basel). 2023. PMID: 37508067 Free PMC article.
-
Genetic Characterization of Small Ruminant Lentiviruses Isolated from Dairy Sheep in Greece.Viruses. 2024 Mar 31;16(4):547. doi: 10.3390/v16040547. Viruses. 2024. PMID: 38675890 Free PMC article.
References
-
- Adams D. S. et al. (1984) ‘Global Survey of Serological Evidence of Caprine Arthritis-Encephalitis Virus Infection’, The Veterinary Record, 115: 493–5. - PubMed
-
- Amills M., Capote J., and Tosser-Klopp G. (2017) ‘Goat Domestication and Breeding: A Jigsaw of Historical, Biological and Molecular Data with Missing Pieces’, Animal Genetics, 48: 631–44. - PubMed
-
- Anonymous (1900) ‘Fat-Tailed Sheep in Central Asia and California’. Pacific Rural Press, 60: 132–3.
LinkOut - more resources
Full Text Sources

