Genomic structural variation: A complex but important driver of human evolution
- PMID: 36794631
- PMCID: PMC10329998
- DOI: 10.1002/ajpa.24713
Genomic structural variation: A complex but important driver of human evolution
Abstract
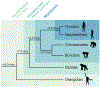
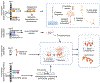
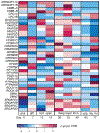
Structural variants (SVs)-including duplications, deletions, and inversions of DNA-can have significant genomic and functional impacts but are technically difficult to identify and assay compared with single-nucleotide variants. With the aid of new genomic technologies, it has become clear that SVs account for significant differences across and within species. This phenomenon is particularly well-documented for humans and other primates due to the wealth of sequence data available. In great apes, SVs affect a larger number of nucleotides than single-nucleotide variants, with many identified SVs exhibiting population and species specificity. In this review, we highlight the importance of SVs in human evolution by (1) how they have shaped great ape genomes resulting in sensitized regions associated with traits and diseases, (2) their impact on gene functions and regulation, which subsequently has played a role in natural selection, and (3) the role of gene duplications in human brain evolution. We further discuss how to incorporate SVs in research, including the strengths and limitations of various genomic approaches. Finally, we propose future considerations in integrating existing data and biospecimens with the ever-expanding SV compendium propelled by biotechnology advancements.
Keywords: brain; gene duplications; genomes; human evolution; structural variation.
© 2023 American Association of Biological Anthropologists.
Figures
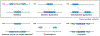



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
