Comparative Genome Analysis of Enterococcus cecorum Reveals Intercontinental Spread of a Lineage of Clinical Poultry Isolates
- PMID: 36794931
- PMCID: PMC10117131
- DOI: 10.1128/msphere.00495-22
Comparative Genome Analysis of Enterococcus cecorum Reveals Intercontinental Spread of a Lineage of Clinical Poultry Isolates
Abstract
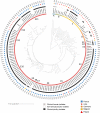
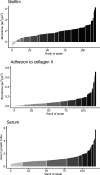
Enterococcus cecorum is an emerging pathogen responsible for osteomyelitis, spondylitis, and femoral head necrosis causing animal suffering and mortality and requiring antimicrobial use in poultry. Paradoxically, E. cecorum is a common inhabitant of the intestinal microbiota of adult chickens. Despite evidence suggesting the existence of clones with pathogenic potential, the genetic and phenotypic relatedness of disease-associated isolates remains little investigated. Here, we sequenced and analyzed the genomes and characterized the phenotypes of more than 100 isolates, the majority of which were collected over the last 10 years from 16 French broiler farms. Comparative genomics, genome-wide association studies, and the measured susceptibility to serum, biofilm-forming capacity, and adhesion to chicken type II collagen were used to identify features associated with clinical isolates. We found that none of the tested phenotypes could discriminate the origin of the isolates or the phylogenetic group. Instead, we found that most clinical isolates are grouped phylogenetically, and our analyses selected six genes that discriminate 94% of isolates associated with disease from those that are not. Analysis of the resistome and the mobilome revealed that multidrug-resistant clones of E. cecorum cluster into a few clades and that integrative conjugative elements and genomic islands are the main carriers of antimicrobial resistance. This comprehensive genomic analysis shows that disease-associated clones of E. cecorum belong mainly to one phylogenetic clade. IMPORTANCE Enterococcus cecorum is an important pathogen of poultry worldwide. It causes a number of locomotor disorders and septicemia, particularly in fast-growing broilers. Animal suffering, antimicrobial use, and associated economic losses require a better understanding of disease-associated E. cecorum isolates. To address this need, we performed whole-genome sequencing and analysis of a large collection of isolates responsible for outbreaks in France. By providing the first data set on the genetic diversity and resistome of E. cecorum strains circulating in France, we pinpoint an epidemic lineage that is probably also circulating elsewhere that should be targeted preferentially by preventive strategies in order to reduce the burden of E. cecorum-related diseases.
Keywords: Enterococcus cecorum; antimicrobial resistance; avian pathogenesis; comparative genomics; poultry; veterinary pathogens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Devriese LA, Dutta GN, Farrow JAE, Vandekerckhove A, Phillips BA. 1983. Streptococcus cecorum, a new species isolated from chickens. Int J Syst Bacteriol 33:772–776. doi:10.1099/00207713-33-4-772. - DOI
-
- Dolka B, Golębiewska-Kosakowska M, Krajewski K, Kwieciński P, Nowak T, Zubstarski J, Wilczyński J, Szeleszczuk P. 2017. Occurrence of Enterococcus spp. in poultry in Poland based on 2014–2015 data. Med Weter 73:220–224.
-
- Souillard R, Allain V, Toux J-Y, Lecaer V, Lahmar A, Tatone F, Amenna-Bernard A, Le Bouquin S. 2019. Synthèse des pathologies aviaires observées en 2018 par le Réseau National d’Observations Épidémiologiques en Aviculture (RNOEA). Bull Epidemiol Sante Anim 88:1–12.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
