Antimicrobial resistance patterns and characterisation of emerging beta-lactamase-producing Escherichia coli in camels sampled from Northern Kenya
- PMID: 36795022
- PMCID: PMC10188057
- DOI: 10.1002/vms3.1090
Antimicrobial resistance patterns and characterisation of emerging beta-lactamase-producing Escherichia coli in camels sampled from Northern Kenya
Abstract
Background: Animal husbandry practices in different livestock production systems and increased livestock-wildlife interactions are thought to be primary drivers of antimicrobial resistance (AMR) in Arid and Semi-Arid Lands (ASALs). Despite a tenfold increase in the camel population within the last decade, paired with widespread use of camel products, there is a lack of comprehensive information concerning beta-lactamase-producing Escherichia coli (E. coli) within these production systems.
Objectives: Our study sought to establish an AMR profile and to identify and characterise emerging beta-lactamase-producing E. coli isolated from faecal samples obtained from camel herds in Northern Kenya.
Methods: The antimicrobial susceptibility profiles of E. coli isolates were established using the disk diffusion method, with beta-lactamase (bla) gene PCR product sequencing performed for phylogenetic grouping and genetic diversity assessments.
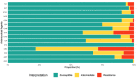
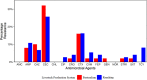
Results: Here we show, among the recovered E. coli isolates (n = 123), the highest level of resistance was observed for cefaclor at 28.5% of isolates, followed by cefotaxime at 16.3% and ampicillin at 9.7%. Moreover, extended-spectrum beta-lactamase (ESBL)-producing E. coli harbouring the blaCTX-M-15 or blaCTX-M-27 genes were detected in 3.3% of total samples, and are associated with phylogenetic groups B1, B2 and D. Multiple variants of non-ESBL blaTEM genes were detected, the majority of which were the blaTEM-1 and blaTEM-116 genes.
Conclusions: Findings from this study shed light on the increased occurrence of ESBL- and non-ESBL-encoding gene variants in E. coli isolates with demonstrated multidrug resistant phenotypes. This study highlights the need for an expanded One Health approach to understanding AMR transmission dynamics, drivers of AMR development, and appropriate practices for antimicrobial stewardship in camel production systems within ASALs.
Keywords: antimicrobial resistance; beta-lactamase-producing Escherichia coli; camels; livestock production systems.
© 2023 The Authors. Veterinary Medicine and Science published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts to declare.
Figures
References
-
- Aliwa, B. O. , & Kaindi, D. W. M. (2019). Antibiotic resistance of Clostridium perfringens isolated from raw camel milk in Isiolo County, Kenya. Annals of Applied Microbiology & Biotechnology Journal, 3(1), 3–7.
-
- Awosile, B. , McClure, J. , Sanchez, J. , Rodriguez‐Lecompte, J. C. , Keefe, G. , & Heider, L. C. (2018). Salmonella enterica and extended‐spectrum cephalosporin‐resistant Escherichia coli recovered from Holstein dairy calves from 8 farms in New Brunswick, Canada. Journal of Dairy Science, 101(4), 3271–3284. 10.3168/JDS.2017-13277 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

