EGFR inhibition leads to enhanced desmosome assembly and cardiomyocyte cohesion via ROCK activation
- PMID: 36795511
- PMCID: PMC10070108
- DOI: 10.1172/jci.insight.163763
EGFR inhibition leads to enhanced desmosome assembly and cardiomyocyte cohesion via ROCK activation
Abstract
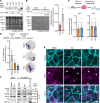
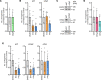
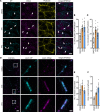
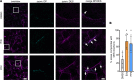
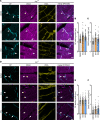
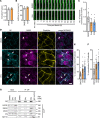
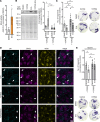
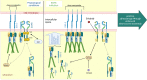
Arrhythmogenic cardiomyopathy (AC) is a familial heart disease partly caused by impaired desmosome turnover. Thus, stabilization of desmosome integrity may provide new treatment options. Desmosomes, apart from cellular cohesion, provide the structural framework of a signaling hub. Here, we investigated the role of the epidermal growth factor receptor (EGFR) in cardiomyocyte cohesion. We inhibited EGFR under physiological and pathophysiological conditions using the murine plakoglobin-KO AC model, in which EGFR was upregulated. EGFR inhibition enhanced cardiomyocyte cohesion. Immunoprecipitation showed an interaction of EGFR and desmoglein 2 (DSG2). Immunostaining and atomic force microscopy (AFM) revealed enhanced DSG2 localization and binding at cell borders upon EGFR inhibition. Enhanced area composita length and desmosome assembly were observed upon EGFR inhibition, confirmed by enhanced DSG2 and desmoplakin (DP) recruitment to cell borders. PamGene Kinase assay performed in HL-1 cardiomyocytes treated with erlotinib, an EGFR inhibitor, revealed upregulation of Rho-associated protein kinase (ROCK). Erlotinib-mediated desmosome assembly and cardiomyocyte cohesion were abolished upon ROCK inhibition. Thus, inhibiting EGFR and, thereby, stabilizing desmosome integrity via ROCK might provide treatment options for AC.
Keywords: Cardiology; Cardiovascular disease; Cell Biology; Cell migration/adhesion.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

