A Unique Cellular Organization of Human Distal Airways and Its Disarray in Chronic Obstructive Pulmonary Disease
- PMID: 36796082
- PMCID: PMC10161760
- DOI: 10.1164/rccm.202207-1384OC
A Unique Cellular Organization of Human Distal Airways and Its Disarray in Chronic Obstructive Pulmonary Disease
Abstract
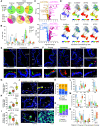
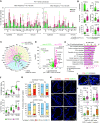
Rationale: Remodeling and loss of distal conducting airways, including preterminal and terminal bronchioles (pre-TBs/TBs), underlie progressive airflow limitation in chronic obstructive pulmonary disease (COPD). The cellular basis of these structural changes remains unknown. Objectives: To identify biological changes in pre-TBs/TBs in COPD at single-cell resolution and determine their cellular origin. Methods: We established a novel method of distal airway dissection and performed single-cell transcriptomic profiling of 111,412 cells isolated from different airway regions of 12 healthy lung donors and pre-TBs of 5 patients with COPD. Imaging CyTOF and immunofluorescence analysis of pre-TBs/TBs from 24 healthy lung donors and 11 subjects with COPD were performed to characterize cellular phenotypes at a tissue level. Region-specific differentiation of basal cells isolated from proximal and distal airways was studied using an air-liquid interface model. Measurements and Main Results: The atlas of cellular heterogeneity along the proximal-distal axis of the human lung was assembled and identified region-specific cellular states, including SCGB3A2+ SFTPB+ terminal airway-enriched secretory cells (TASCs) unique to distal airways. TASCs were lost in COPD pre-TBs/TBs, paralleled by loss of region-specific endothelial capillary cells, increased frequency of CD8+ T cells normally enriched in proximal airways, and augmented IFN-γ signaling. Basal cells residing in pre-TBs/TBs were identified as a cellular origin of TASCs. Regeneration of TASCs by these progenitors was suppressed by IFN-γ. Conclusions: Altered maintenance of the unique cellular organization of pre-TBs/TBs, including loss of the region-specific epithelial differentiation in these bronchioles, represents the cellular manifestation and likely the cellular basis of distal airway remodeling in COPD.
Keywords: bronchiole; epithelium; heterogeneity; regeneration; remodeling.
Figures


Comment in
-
Taking Small Airways in Chronic Obstructive Pulmonary Disease to TASC.Am J Respir Crit Care Med. 2023 May 1;207(9):1114-1115. doi: 10.1164/rccm.202302-0295ED. Am J Respir Crit Care Med. 2023. PMID: 36821491 Free PMC article. No abstract available.
References
-
- Haefeli-Bleuer B, Weibel ER. Morphometry of the human pulmonary acinus. Anat Rec . 1988;220:401–414. - PubMed
-
- Webb WR. Thin-section CT of the secondary pulmonary lobule: anatomy and the image—the 2004 Fleischner Lecture. Radiology . 2006;239:322–338. - PubMed
-
- Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, Buzatu L, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med . 2004;350:2645–2653. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

