Natural variation in gene expression and viral susceptibility revealed by neural progenitor cell villages
- PMID: 36796362
- PMCID: PMC10581885
- DOI: 10.1016/j.stem.2023.01.010
Natural variation in gene expression and viral susceptibility revealed by neural progenitor cell villages
Abstract
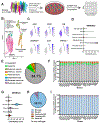
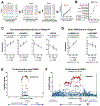
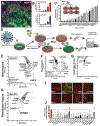
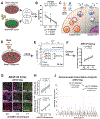
Human genome variation contributes to diversity in neurodevelopmental outcomes and vulnerabilities; recognizing the underlying molecular and cellular mechanisms will require scalable approaches. Here, we describe a "cell village" experimental platform we used to analyze genetic, molecular, and phenotypic heterogeneity across neural progenitor cells from 44 human donors cultured in a shared in vitro environment using algorithms (Dropulation and Census-seq) to assign cells and phenotypes to individual donors. Through rapid induction of human stem cell-derived neural progenitor cells, measurements of natural genetic variation, and CRISPR-Cas9 genetic perturbations, we identified a common variant that regulates antiviral IFITM3 expression and explains most inter-individual variation in susceptibility to the Zika virus. We also detected expression QTLs corresponding to GWAS loci for brain traits and discovered novel disease-relevant regulators of progenitor proliferation and differentiation such as CACHD1. This approach provides scalable ways to elucidate the effects of genes and genetic variation on cellular phenotypes.
Keywords: CACHD1; CRISPR-Cas9 screen; Neurogenin-2; Zika virus; cell villages; neural progenitor cells; neurodevelopmental disorders; proliferation.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.E. is a founder of Q-State Biosciences, Quralis, and Enclear Therapies; an employee and shareholder of BioMarin; and a member of Cell Stem Cell’s advisory board.
Figures



Comment in
-
Pandemic city: Village-in-a-dish unlocks dynamic genetic effects in the brain.Cell Stem Cell. 2023 Mar 2;30(3):239-241. doi: 10.1016/j.stem.2023.02.002. Cell Stem Cell. 2023. PMID: 36868190
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

