Genomic epidemiology of SARS-CoV-2 infections in The Gambia: an analysis of routinely collected surveillance data between March, 2020, and January, 2022
- PMID: 36796985
- PMCID: PMC9928486
- DOI: 10.1016/S2214-109X(22)00553-8
Genomic epidemiology of SARS-CoV-2 infections in The Gambia: an analysis of routinely collected surveillance data between March, 2020, and January, 2022
Abstract
Background: COVID-19, caused by SARS-CoV-2, is one of the deadliest pandemics of the past 100 years. Genomic sequencing has an important role in monitoring of the evolution of the virus, including the detection of new viral variants. We aimed to describe the genomic epidemiology of SARS-CoV-2 infections in The Gambia.
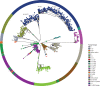
Methods: Nasopharyngeal or oropharyngeal swabs collected from people with suspected cases of COVID-19 and international travellers were tested for SARS-CoV-2 with standard RT-PCR methods. SARS-CoV-2-positive samples were sequenced according to standard library preparation and sequencing protocols. Bioinformatic analysis was done using ARTIC pipelines and Pangolin was used to assign lineages. To construct phylogenetic trees, sequences were first stratified into different COVID-19 waves (waves 1-4) and aligned. Clustering analysis was done and phylogenetic trees constructed.
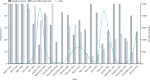
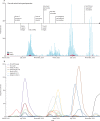
Findings: Between March, 2020, and January, 2022, 11 911 confirmed cases of COVID-19 were recorded in The Gambia, and 1638 SARS-CoV-2 genomes were sequenced. Cases were broadly distributed into four waves, with more cases during the waves that coincided with the rainy season (July-October). Each wave occurred after the introduction of new viral variants or lineages, or both, generally those already established in Europe or in other African countries. Local transmission was higher during the first and third waves (ie, those that corresponded with the rainy season), in which the B.1.416 lineage and delta (AY.34.1) were dominant, respectively. The second wave was driven by the alpha and eta variants and the B.1.1.420 lineage. The fourth wave was driven by the omicron variant and was predominantly associated with the BA.1.1 lineage.
Interpretation: More cases of SARS-CoV-2 infection were recorded in The Gambia during peaks of the pandemic that coincided with the rainy season, in line with transmission patterns for other respiratory viruses. The introduction of new lineages or variants preceded epidemic waves, highlighting the importance of implementing well structured genomic surveillance at a national level to detect and monitor emerging and circulating variants.
Funding: Medical Research Unit The Gambia at London School of Hygiene & Tropical Medicine, UK Research and Innovation, WHO.
Copyright © 2023 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests We declare no competing interests.
Figures



References
-
- Worldometer COVID-19 live coronavirus pandemic. 2022. https://www.worldometers.info/coronavirus/
-
- Chen J, Lu H. New challenges to fighting COVID-19: virus variants, potential vaccines, and development of antivirals. Biosci Trends. 2021;15:126–128. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

