Tandemly duplicated CYP82Ds catalyze 14-hydroxylation in triptolide biosynthesis and precursor production in Saccharomyces cerevisiae
- PMID: 36797237
- PMCID: PMC9936527
- DOI: 10.1038/s41467-023-36353-y
Tandemly duplicated CYP82Ds catalyze 14-hydroxylation in triptolide biosynthesis and precursor production in Saccharomyces cerevisiae
Abstract
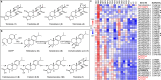
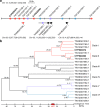
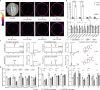
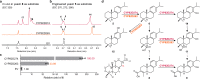
Triptolide is a valuable multipotent antitumor diterpenoid in Tripterygium wilfordii, and its C-14 hydroxyl group is often selected for modification to enhance both the bioavailability and antitumor efficacy. However, the mechanism for 14-hydroxylation formation remains unknown. Here, we discover 133 kb of tandem duplicated CYP82Ds encoding 11 genes on chromosome 12 and characterize CYP82D274 and CYP82D263 as 14-hydroxylases that catalyze the metabolic grid in triptolide biosynthesis. The two CYP82Ds catalyze the aromatization of miltiradiene, which has been repeatedly reported to be a spontaneous process. In vivo assays and evaluations of the kinetic parameters of CYP82Ds indicate the most significant affinity to dehydroabietic acid among multiple intermediates. The precursor 14-hydroxy-dehydroabietic acid is successfully produced by engineered Saccharomyces cerevisiae. Our study provides genetic elements for further elucidation of the downstream biosynthetic pathways and heterologous production of triptolide and of the currently intractable biosynthesis of other 14-hydroxyl labdane-type secondary metabolites.
© 2023. The Author(s).
Conflict of interest statement
Yi.Z., J.G., L.M., and W.G. are inventors of Chinese invention patents (application No. CN202210059108.2 and CN202210059174.X) related to the cytochrome P450 enzymes described in the paper. Other authors declare no competing interests.
Figures


Similar articles
-
Production of Putative Diterpene Carboxylic Acid Intermediates of Triptolide in Yeast.Molecules. 2017 Jun 13;22(6):981. doi: 10.3390/molecules22060981. Molecules. 2017. PMID: 28608823 Free PMC article.
-
Identification of geranylgeranyl diphosphate synthase genes from Tripterygium wilfordii.Plant Cell Rep. 2015 Dec;34(12):2179-88. doi: 10.1007/s00299-015-1860-3. Epub 2015 Oct 8. Plant Cell Rep. 2015. PMID: 26449416
-
Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis.Nat Commun. 2020 Feb 20;11(1):971. doi: 10.1038/s41467-020-14776-1. Nat Commun. 2020. PMID: 32080175 Free PMC article.
-
Triptolide: pharmacological spectrum, biosynthesis, chemical synthesis and derivatives.Theranostics. 2021 May 24;11(15):7199-7221. doi: 10.7150/thno.57745. eCollection 2021. Theranostics. 2021. PMID: 34158845 Free PMC article. Review.
-
Therapeutic targets of thunder god vine (Tripterygium wilfordii hook) in rheumatoid arthritis (Review).Mol Med Rep. 2020 Jun;21(6):2303-2310. doi: 10.3892/mmr.2020.11052. Epub 2020 Apr 2. Mol Med Rep. 2020. PMID: 32323812 Review.
Cited by
-
Targeted metabolic engineering of key biosynthetic genes improves triptolide production in Tripterygium wilfordii hairy roots.Plant Cell Rep. 2025 May 22;44(6):129. doi: 10.1007/s00299-025-03518-6. Plant Cell Rep. 2025. PMID: 40404876
-
Functional Study and Efficient Catalytic Element Mining of CYP76AHs in Salvia Plants.Molecules. 2023 Jun 12;28(12):4711. doi: 10.3390/molecules28124711. Molecules. 2023. PMID: 37375266 Free PMC article.
-
Integrated transcriptomics and metabolomics provide insights into the biosynthesis of militarine in the cell suspension culture system of Bletilla striata.Adv Biotechnol (Singap). 2024 Jul 16;2(3):25. doi: 10.1007/s44307-024-00032-w. Adv Biotechnol (Singap). 2024. PMID: 39883253 Free PMC article.
-
Divergent multifunctional P450s-empowered biosynthesis of bioactive tripterifordin and cryptic atiserenoids in Aconitum implies convergent evolution.Nat Commun. 2025 Jul 1;16(1):5857. doi: 10.1038/s41467-025-61188-0. Nat Commun. 2025. PMID: 40595667 Free PMC article.
-
CYP72D19 from Tripterygium wilfordii catalyzes C-2 hydroxylation of abietane-type diterpenoids.Plant Cell Rep. 2023 Nov;42(11):1733-1744. doi: 10.1007/s00299-023-03059-w. Epub 2023 Aug 24. Plant Cell Rep. 2023. PMID: 37615706
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

