The ULK3 kinase is a determinant of keratinocyte self-renewal and tumorigenesis targeting the arginine methylome
- PMID: 36797248
- PMCID: PMC9935893
- DOI: 10.1038/s41467-023-36410-6
The ULK3 kinase is a determinant of keratinocyte self-renewal and tumorigenesis targeting the arginine methylome
Abstract
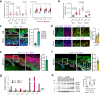
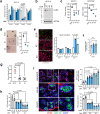
Epigenetic mechanisms oversee epidermal homeostasis and oncogenesis. The identification of kinases controlling these processes has direct therapeutic implications. We show that ULK3 is a nuclear kinase with elevated expression levels in squamous cell carcinomas (SCCs) arising in multiple body sites, including skin and Head/Neck. ULK3 loss by gene silencing or deletion reduces proliferation and clonogenicity of human keratinocytes and SCC-derived cells and affects transcription impinging on stem cell-related and metabolism programs. Mechanistically, ULK3 directly binds and regulates the activity of two histone arginine methyltransferases, PRMT1 and PRMT5 (PRMT1/5), with ULK3 loss compromising PRMT1/5 chromatin association to specific genes and overall methylation of histone H4, a shared target of these enzymes. These findings are of translational significance, as downmodulating ULK3 by RNA interference or locked antisense nucleic acids (LNAs) blunts the proliferation and tumorigenic potential of SCC cells and promotes differentiation in two orthotopic models of skin cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

