Oxidized mitochondrial DNA induces gasdermin D oligomerization in systemic lupus erythematosus
- PMID: 36797275
- PMCID: PMC9935630
- DOI: 10.1038/s41467-023-36522-z
Oxidized mitochondrial DNA induces gasdermin D oligomerization in systemic lupus erythematosus
Abstract
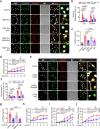
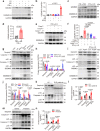
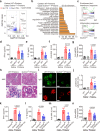
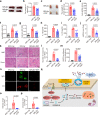
Although extracellular DNA is known to form immune complexes (ICs) with autoantibodies in systemic lupus erythematosus (SLE), the mechanisms leading to the release of DNA from cells remain poorly characterized. Here, we show that the pore-forming protein, gasdermin D (GSDMD), is required for nuclear DNA and mitochondrial DNA (mtDNA) release from neutrophils and lytic cell death following ex vivo stimulation with serum from patients with SLE and IFN-γ. Mechanistically, the activation of FcγR downregulated Serpinb1 following ex vivo stimulation with serum from patients with SLE, leading to spontaneous activation of both caspase-1/caspase-11 and cleavage of GSDMD into GSDMD-N. Furthermore, mtDNA oxidization promoted GSDMD-N oligomerization and cell death. In addition, GSDMD, but not peptidyl arginine deiminase 4 is necessary for extracellular mtDNA release from low-density granulocytes from SLE patients or healthy human neutrophils following incubation with ICs. Using the pristane-induced lupus model, we show that disease severity is significantly reduced in mice with neutrophil-specific Gsdmd deficiency or following treatment with the GSDMD inhibitor, disulfiram. Altogether, our study highlights an important role for oxidized mtDNA in inducing GSDMD oligomerization and pore formation. These findings also suggest that GSDMD might represent a possible therapeutic target in SLE.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Effects of Gasdermin D in Modulating Murine Lupus and its Associated Organ Damage.Arthritis Rheumatol. 2020 Dec;72(12):2118-2129. doi: 10.1002/art.41444. Epub 2020 Oct 17. Arthritis Rheumatol. 2020. PMID: 32692482 Free PMC article.
-
Novel Effects of Combination Therapy Through Inhibition of Caspase-1/Gasdermin D Induced-Pyroptosis in Lupus Nephritis.Front Immunol. 2021 Nov 19;12:720877. doi: 10.3389/fimmu.2021.720877. eCollection 2021. Front Immunol. 2021. PMID: 34867948 Free PMC article.
-
Neutrophil Extracellular Trap Mitochondrial DNA and Its Autoantibody in Systemic Lupus Erythematosus and a Proof-of-Concept Trial of Metformin.Arthritis Rheumatol. 2015 Dec;67(12):3190-200. doi: 10.1002/art.39296. Arthritis Rheumatol. 2015. PMID: 26245802 Clinical Trial.
-
Gasdermins and their role in immunity and inflammation.J Exp Med. 2019 Nov 4;216(11):2453-2465. doi: 10.1084/jem.20190545. Epub 2019 Sep 23. J Exp Med. 2019. PMID: 31548300 Free PMC article. Review.
-
Regulation of Lytic and Non-Lytic Functions of Gasdermin Pores.J Mol Biol. 2022 Feb 28;434(4):167246. doi: 10.1016/j.jmb.2021.167246. Epub 2021 Sep 17. J Mol Biol. 2022. PMID: 34537232 Review.
Cited by
-
New Mechanisms and Therapeutic Targets in Systemic Lupus Erythematosus.MedComm (2020). 2025 Jun 9;6(6):e70246. doi: 10.1002/mco2.70246. eCollection 2025 Jun. MedComm (2020). 2025. PMID: 40491969 Free PMC article. Review.
-
Caspase-11/GSDMD contributes to the progression of hyperuricemic nephropathy by promoting NETs formation.Cell Mol Life Sci. 2024 Mar 4;81(1):114. doi: 10.1007/s00018-024-05136-z. Cell Mol Life Sci. 2024. PMID: 38436813 Free PMC article.
-
New insights into constitutive neutrophil death.Cell Death Discov. 2025 Jan 12;11(1):6. doi: 10.1038/s41420-025-02287-1. Cell Death Discov. 2025. PMID: 39800780 Free PMC article. Review.
-
Formation of neutrophil extracellular traps in the early stages exacerbate the healing process by regulating macrophage polarization in Achilles tendon-bone injury.J Tissue Eng. 2025 Aug 19;16:20417314251348038. doi: 10.1177/20417314251348038. eCollection 2025 Jan-Dec. J Tissue Eng. 2025. PMID: 40843411 Free PMC article.
-
Precise Orchestration of Gasdermins' Pore-Forming Function by Posttranslational Modifications in Health and Disease.Int J Biol Sci. 2023 Sep 18;19(15):4931-4947. doi: 10.7150/ijbs.86869. eCollection 2023. Int J Biol Sci. 2023. PMID: 37781519 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

