Site-specific encoding of photoactivity and photoreactivity into antibody fragments
- PMID: 36797401
- PMCID: PMC10229432
- DOI: 10.1038/s41589-022-01251-9
Site-specific encoding of photoactivity and photoreactivity into antibody fragments
Abstract
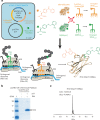
Design of biomolecules that perform two or more distinct functions in response to light remains challenging. Here, we have introduced concurrent photoactivity and photoreactivity into an epidermal growth factor receptor (EGFR)-targeting antibody fragment, 7D12. This was achieved by site-specific incorporation of photocaged tyrosine (pcY) for photoactivity and p-benzoyl-ʟ-phenylalanine (Bpa) for photoreactivity into 7D12. We identified a position for installing Bpa in 7D12 that has minimal effect on 7D12-EGFR binding affinity in the absence of light. Upon exposure to 365-nm light, this Bpa-containing 7D12 mutant forms a covalent bond with EGFR in an antigen-specific manner. We then developed a method for site-specific incorporation of pcY and Bpa at two distinct sites in 7D12. Finally, we demonstrated that in the absence of light, this pcY- and Bpa-containing mutant of 7D12 does not bind to EGFR, but irradiation with 365-nm light activates (1) specific binding and (2) covalent bond formation with EGFR.
© 2023. The Author(s).
Conflict of interest statement
T.B., U.W. and A.S. have filed a patent application related to the research described in this manuscript (Patent application no. GB 2213242.7). The other authors declare no competing interests.
Figures














References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

